













| General Information: |
|
| Name(s) found: |
cfi-1 /
CE31068
[WormBase]
|
| Description(s) found:
Found 22 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 467 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA intracellular [IEA |
| Biological Process: |
neuron differentiation
[IMP cell fate commitment [IMP |
| Molecular Function: |
DNA binding
[IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSVRIDEPQL FVSMSKEPTQ ETVNVGGHHD DSSSNCDERV DDQTEEQKSP PASPDLTANL 60
61 NVFDLESRQK VVQRLLNSQL NLSNLRAPLN LPPIFQALQG PFSIQQQLLG LASGLTAISP 120
121 GLDDYDEENT NQGEPEDLTL GGFRKETSVK SEEPSESGIN ASGPAWSYEE QFKQLYELSD 180
181 DVKRKEWLDD WLNFMHRIGK PVTRIPIMAK QVLDLYELYR LVVQHGGLVE IINKKLWREI 240
241 TKGLNLPSSI TSAAFTLRTQ YQKYLYDYEC EKEKLSNQSD LQQAIDGNRR EAPGRRTAPS 300
301 FPLPFQLPHA ASAAATMLNN QLNGLGMRND LLDDENTLSL QASGLFGTSY GAEQMAILEA 360
361 HQRNLERAQR AVQQQVARQS LGLTACSNGN GGNIHNSGRE STSSNDSDIP AKRPKLENDV 420
421 KTNGASSMRI STKHSDNSKT SMSVSMEING ITYQGVLFAL DETVSES |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
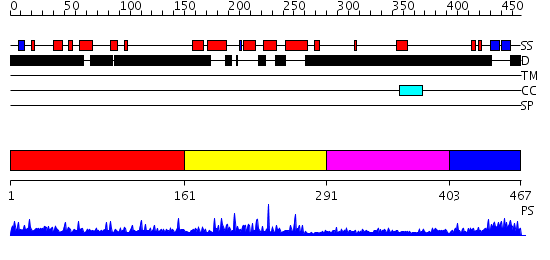
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..160] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [161..290] | 36.09691 | DNA-binding domain from the dead ringer protein | |
| 3 | View Details | [291..402] | 1.02 | Structure of the ribosomal 80S-eEF2-sordarin complex from yeast obtained by docking atomic models for RNA and protein components into a 11.7 A cryo-EM map. This file, 1S1I, Contains 60S subunit. The 40S Ribosomal Subunit Is In File 1S1H. | |
| 4 | View Details | [403..467] | 2.103995 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||
| 2 |
|
||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)