













| General Information: |
|
| Name(s) found: |
aak-2 /
CE31223
[WormBase]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Caenorhabditis elegans |
| Length: | 626 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IEA |
| Biological Process: |
signal transduction
[IEA determination of adult lifespan [IMP protein amino acid phosphorylation [IDA |
| Molecular Function: |
magnesium ion binding
[IEA ATP binding [IEA protein tyrosine kinase activity [IEA zinc ion binding [IEA non-membrane spanning protein tyrosine kinase activity [IEA protein kinase activity [IDA protein serine/threonine kinase activity [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MFSHQDRDRD RKEDGGGDGT EMKSKSRSQP SGLNRVKNLS RKLSAKSRKE RKDRDSTDNS 60
61 SKMSSPGGET STKQQQELKA QIKIGHYILK ETLGVGTFGK VKVGIHETTQ YKVAVKILNR 120
121 QKIKSLDVVG KIRREIQNLS LFRHPHIIRL YQVISTPSDI FMIMEHVSGG ELFDYIVKHG 180
181 RLKTAEARRF FQQIISGVDY CHRHMVVHRD LKPENLLLDE QNNVKIADFG LSNIMTDGDF 240
241 LRTSCGSPNY AAPEVISGKL YAGPEVDVWS CGVILYALLC GTLPFDDEHV PSLFRKIKSG 300
301 VFPTPDFLER PIVNLLHHML CVDPMKRATI KDVIAHEWFQ KDLPNYLFPP INESEASIVD 360
361 IEAVREVTEF QRYHVAEEEV TSALLGDDPH HHLSIAYNLI VDNKRIADET AKLSIEEFYQ 420
421 VTPNKGPGPV HRHPERIAAS VSSKITPTLD NTEASGANRN KRAKWHLGIR SQSRPEDIMF 480
481 EVFRAMKQLD MEWKVLNPYH VIVRRKPDAP AADPPKMSLQ LYQVDQRSYL LDFKSLADEE 540
541 SGSASASSSR HASMSMPQKP AGIRGTRTSS MPQAMSMEAS IEKMEVHDFS DMSCDVTPPP 600
601 SPGGAKLSQT MQFFEICAAL IGTLAR |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
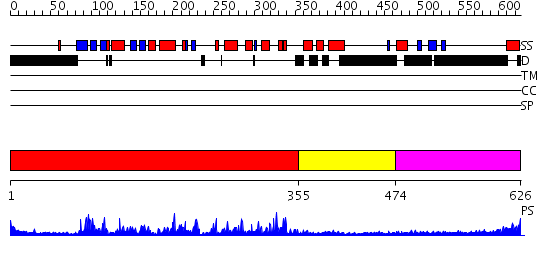
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..354] | 81.0 | No description for 2h6dA was found. | |
| 2 | View Details | [355..473] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [474..626] | 17.69897 | No description for 2v8qA was found. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)