













| General Information: |
|
| Name(s) found: |
CE30792
[WormBase]
|
| Description(s) found:
Found 18 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 683 amino acids |
Gene Ontology: |
|
| Cellular Component: |
integral to membrane
[IEA |
| Biological Process: |
lipid metabolic process
[IEA |
| Molecular Function: |
triglyceride lipase activity
[IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MPSLVAFNRQ WNIGSDDFVF PEITEALVRL SWVAFGVSVF VFHFPLACST HNLTLQLLGL 60
61 LVINVITILL AILTAVISAR GTIMNSQPRK LIPRLLYIRL PIFIAEIALT ILATIHAFKS 120
121 TNGGEGCDFA TIARVTIILE WFLIFTAFAG LVIVFHLTEN ELEDSASIAQ KSWSRRMRIF 180
181 KIGQDHSMRV ALDDLAILIS SFFVDSDLVP SDVVAGLLLA YHSPNNQYPP VKDKEHESVP 240
241 KWMNLKDAEY FLHHATCVYG WPTYILYNCG LKSIYRLFRK LQCCGRMRCA QSPCVVQQQR 300
301 RERIELTELS NGKLVKKGKS MVVEDNCCYC NTAAVVLANE ARNIDLQFMS FRNRVYEVPF 360
361 AVIADHDKKS IVITIRGSCS LIDLVTDLSL EDELMTVDVD QDATLREDEE IDKRGDVRVH 420
421 RGMLRSARYV FDTLNKNKIL NDLFISNPSY QLVVCGHSLG AGVGSLLTML LKQEYPSVIC 480
481 YAFAPPGCVI SEFGQDEMEK YVMSVVSGDD IVSRMSFQSL HRLRERVFQE LLACQRAKYE 540
541 ILIRGVYQLF FKYPWGDELS GVPRISASPD SPTTPDIESA LLTRRNSYGA SDPTSEASPT 600
601 PPQRNPNHNK RLQLYVPGRV IYLSADQDTG ATVETWIDPK CLSDVKLSVS VLSDHLPAAV 660
661 QKFLNSAAIT FEQREEIVSS QPV |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
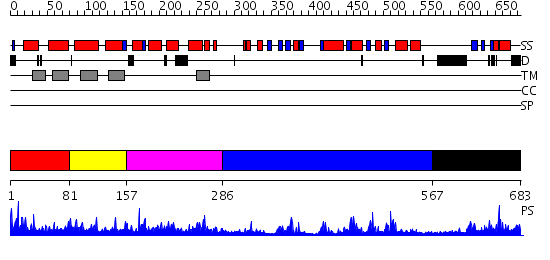
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..80] | 3.026986 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [81..156] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [157..285] | 1.152973 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [286..566] | 48.69897 | Triacylglycerol lipase | |
| 5 | View Details | [567..683] | 1.239985 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||
| 1 | No functions predicted. | |||||||||
| 2 | No functions predicted. | |||||||||
| 3 | No functions predicted. | |||||||||
| 4 |
|
|||||||||
| 5 | No functions predicted. |
| Source: Reynolds et al. 2008. Manuscript submitted |

|