













| General Information: |
|
| Name(s) found: |
CE31479
[WormBase]
|
| Description(s) found:
Found 19 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 484 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IEA |
| Biological Process: |
protein modification process
[IEA nucleotide-excision repair [IEA |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MATESALIKV HVKSPSNKYD VEIAADASVS ELKDKVLVFV PTANKEQVCI IYTGKILKDE 60
61 ETLTQHKIAD GHTVHLVIRN QARPTPAPAA ATPTASSAPT MGGMGSPADI LNNPDAMRSV 120
121 MDNPITQQLL GNPEFMRTII QSNPQFQALI ERNPEVGHIL NDPNVMRQTM EMIRNPNMFQ 180
181 EMMRNHDQAI RNLQGIPGGE AALERLYNDV QEPLLNSATN SLSGNPFASL RGDQSSEPRV 240
241 DRAGQENNEA LPNPWASNAN QATNNQSNNR SADFNSLLDS PGISSLMEQM MSNPSMQASM 300
301 FSPEVINSIR QNMSNNPGLI DSIVGQIPSA RDNPQISEGI RRSFPQMLNM MSDPSVMEAM 360
361 RNPRVSEAFR QIQEGFSTLR REAPQLLNLF QAGAMGGGAF GSDANASSAG ANSANGLADL 420
421 FNSMNMGGGR PSSTAAPVNP EQTYASQLEQ LQSMGFSDRA RNVAALTATF GDLNAAVERL 480
481 LNSP |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
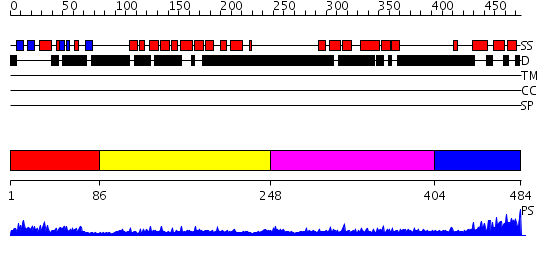
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..85] | 16.30103 | Solution Structure of the N-terminal Ubiquitin-like Domain in the Human Ubiquilin 3 (UBQLN3) | |
| 2 | View Details | [86..247] | 1.33 | No description for 2qsfX was found. | |
| 3 | View Details | [248..403] | 5.39794 | Sec24 | |
| 4 | View Details | [404..484] | 9.0 | Solution Structure of RSGI RUH-016, a UBA Domain from mouse cDNA |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||
| 4 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)