













| General Information: |
|
| Name(s) found: |
hlh-1 /
CE31766
[WormBase]
|
| Description(s) found:
Found 21 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 320 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA |
| Biological Process: |
muscle organ development
[ISS locomotory behavior [IMP nematode larval development [IMP embryonic development ending in birth or egg hatching [IMP positive regulation of growth rate [IMP post-embryonic body morphogenesis [IMP protein heterooligomerization [ISS regulation of transcription [IEA regulation of transcription from RNA polymerase II promoter [ISS |
| Molecular Function: |
transcription regulator activity
[IEA RNA polymerase II transcription factor activity, enhancer binding [ISS bHLH transcription factor binding [ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MNTETSTQSA PSDTYDTSIY YNSSPRVTAN DITTLTSFAA PAPQVLDYAN TQYDIYRNQP 60
61 AYYLPSYAPT APTTFYSDFA NFNVTRSQDF ASVPAVANSS DVKPIIIKQE KSTPNATELI 120
121 IQSRVDSQHE DTTTSTAGGA GVGGPRRTKL DRRKAATMRE RRRLRKVNEA FEVVKQRTCP 180
181 NPNQRLPKVE ILRSAIDYIN NLERMLQQAG KMTKIMEQNQ HLQMTQQING APPHDYVTSS 240
241 HFASSSYNPE NMFDDDDLTD SDDDRDHHKL GNAVDLRRRN SLDRLSRIVA SIPNEEAMTD 300
301 EQLLQPANDV IDGEKKLEML |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
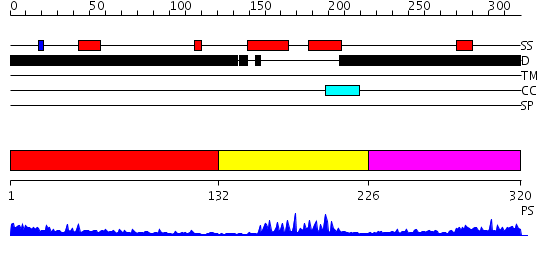
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..131] | 15.268995 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [132..225] | 16.39794 | Myod B/HLH domain | |
| 3 | View Details | [226..320] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)