













| General Information: |
|
| Name(s) found: |
aco-2 /
CE30144
[WormBase]
|
| Description(s) found:
Found 20 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 665 amino acids |
Gene Ontology: |
|
| Cellular Component: |
3-isopropylmalate dehydratase complex
[IEA |
| Biological Process: |
lysine biosynthetic process
[IEA locomotory behavior [IMP hermaphrodite genitalia development [IMP nematode larval development [IMP embryonic development ending in birth or egg hatching [IMP metabolic process [IEA leucine biosynthetic process [IEA tricarboxylic acid cycle [IEA cellular amino acid biosynthetic process [IEA reproduction [IMP growth [IMP |
| Molecular Function: |
hydro-lyase activity
[IEA homoaconitate hydratase activity [IEA 4 iron, 4 sulfur cluster binding [IEA 3-isopropylmalate dehydratase activity [IEA aconitate hydratase activity [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MNSLLRLSHL AGPAHYRALH SSSSIWSKVA ISKFEPKSYL PYEKLSQTVK IVKDRLKRPL 60
61 TLSEKILYGH LDQPKTQDIE RGVSYLRLRP DRVAMQDAVD VMADIPWELK CPKVIGIKLT 120
121 GKLNGWTSAK DVILKVADIL TVKGGTGAIV EYFGPGVDSI SATGMGTICN MGAEIGATTS 180
181 VFPYNESMYK YLEATGRKEI AEEARKYKDL LTADDGANYD QIIEINLDTL TPHVNGPFTP 240
241 DLASSIDKLG ENAKKNGWPL DVKVSLIGSC TNSSYEDMTR AASIAKQALD KGLKAKTIFT 300
301 ITPGSEQVRA TIERDGLSKI FADFGGMVLA NACGPCIGQW DRQDVKKGEK NTIVTSYNRN 360
361 FTGRNDANPA THGFVTSPDI TTAMAISGRL DFNPLTDELT AADGSKFKLQ APTGLDLPPK 420
421 GYDPGEDTFQ APSGSGQVDV SPSSDRLQLL SPFDKWDGKD LEDMKILIKV TGKCTTDHIS 480
481 AAGPWLKYRG HLDNISNNLF LTAINADNGE MNKVKNQVTG EYGAVPATAR KYKADGVRWV 540
541 AIGDENYGEG SSREHAALEP RHLGGRAIIV KSFARIHETN LKKQGMLPLT FANPADYDKI 600
601 DPSDNVSIVG LSSFAPGKPL TAIFKKTNGS KVEVTLNHTF NEQQIEWFKA GSALNRMKEV 660
661 FAKSK |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
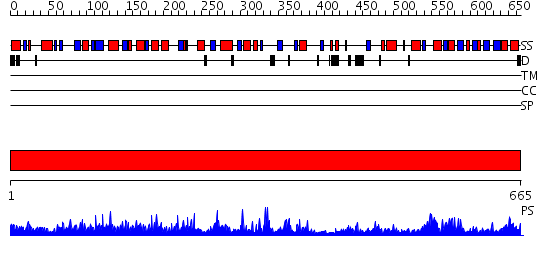
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..665] | 1000.0 | STRUCTURE OF ACTIVATED ACONITASE. FORMATION OF THE (4FE-4S) CLUSTER IN THE CRYSTAL |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.87 |
Source: Reynolds et al. (2008)