













| General Information: |
|
| Name(s) found: |
cku-70 /
CE28984
[WormBase]
|
| Description(s) found:
Found 13 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 691 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IEA DNA-dependent protein kinase-DNA ligase 4 complex [IEA |
| Biological Process: |
determination of adult lifespan
[IMP double-strand break repair via nonhomologous end joining [IEA |
| Molecular Function: |
DNA binding
[IEA ATP-dependent DNA helicase activity [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDETRFEDDD FEGSTANKKY TFFIIDGNPA MFETSKAGEP PEFKLALKLI LDEIVRVCCS 60
61 RSLNNHIGVI VTSTKNSETE GLENSTLLVP MGVLGQEEVN KIKEIAEEEN LLSAVNNYGG 120
121 DHHKSDLSNV LNYCKRVFAS CSNTRHQSVI YLTNNRNPFE RDDFLESSHF KRTKAAVTKI 180
181 IGEGHRKTLG EFSVIMLPED GYKPQDKDEK RKEPWYDLDT EVFSTECDAA ARIRQKITAQ 240
241 RSHATLTVNV GPGVTFDVSV FSMVMEAKPL DHSQKYTRDT EEKIVKTSGY VKKESKMELE 300
301 STEIETQDSV LDETQKMLIR CKNFFCCENS EKNEFLEDSI RNRRDLKKSI EIGGEKIILD 360
361 GDQYEYMNEV NSKGVDFVGF CSMSRVDRET SVVSSKIIQP NDQTTLGSTA IYRTFLDRCW 420
421 ARQQAIVCKY QSRSKQKMRL MALVPFKKDM TLIEKRHENG EDDDDMEDKK PDLLRLEQQR 480
481 AQADSSEWLH EGFMLVGQPF REELRDDFKR FEEQQNVLTE PSTEEQVNTM KQFVKRLTMS 540
541 YNPSFYENPR LLSERSALCL EATGEELIER RDTLEPYYQI PKRLQRVGTE IEKIVETFAL 600
601 NDEIKETKPK ASAGRKRKTD DDEPSTSKKA TMTIRDCIAA DKIGKFKKDE LLSMAVEHCE 660
661 ASKSLKSKTK NDIIAVIEAY LDENPSATWN I |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | hcp1 IP from december2004 | Cheeseman, I.M., et al. (2005) | |
| View Run | hcp2 ip from december | Cheeseman, I.M., et al. (2005) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
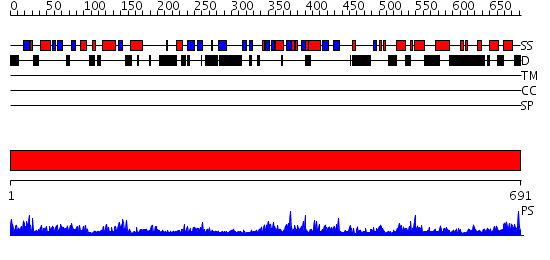
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..691] | 138.0 | DNA binding C-terminal domain of ku70; Ku70 subunit |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)