













| General Information: |
|
| Name(s) found: |
nhr-49 /
CE28943
[WormBase]
|
| Description(s) found:
Found 18 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 476 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IEA |
| Biological Process: |
locomotory behavior
[IMP nematode larval development [IMP positive regulation of growth rate [IMP embryonic development ending in birth or egg hatching [IMP regulation of transcription, DNA-dependent [IEA positive regulation of locomotion [IMP |
| Molecular Function: |
DNA binding
[IEA transcription factor activity [IEA zinc ion binding [IEA steroid hormone receptor activity [IEA sequence-specific DNA binding [IEA steroid binding [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDLVDPLAEP CAVCGDKSTG THYGVISCNG CKGFFRRTVL RDQKFTCRFN KRCVIDKNFR 60
61 CACRYCRFQK CVQVGMKREA IQFERDPVGS PTSGASLNGT PFKKDRSPGY ENGNSNGVGS 120
121 NGMGQENMRT VPQSSSVIDA LMEMEARVNQ EMCNRYRRSQ IFANGSGGSN GNDTDIQQGS 180
181 DSGASAFAPP NRPCTTEDLN EISRTTLLLM VEWAKTINPF MDLSMEDKII LLKNYAPQHL 240
241 ILMPAFRSPD TTRVCLFNNT YMTRDNNTDL NGFAAFKTSN ITPRVLDEIV WPMRQLQMRE 300
301 QEFVCLKALA FLHPEAKGLS NSSQIMIRDA RNRVLKALYA FILDQMPDDA PTRYGNILLL 360
361 APALKALTQL LIENMTLTKF FGLAEVDSLL SEFILDDIND HSTAPVSLQQ HLSSPTTLPT 420
421 NGVSPLNPAG SVGSVSSVSG ITPTGMLSAT LAAPLAIHPL QSQDSILNSE QNNHML |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
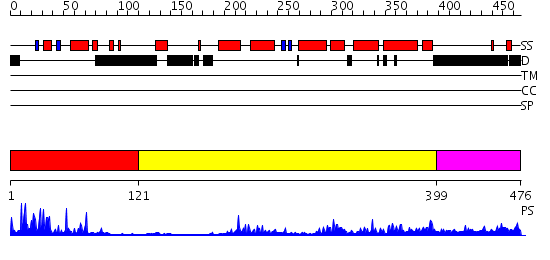
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..120] | 27.39794 | Human Liver Receptor Homologue DNA-Binding Domain (hLRH-1 DBD) in Complex with dsDNA from the hCYP7A1 Promoter | |
| 2 | View Details | [121..398] | 48.154902 | No description for 2gl8A was found. | |
| 3 | View Details | [399..476] | 1.190997 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.86 |
Source: Reynolds et al. (2008)