













| General Information: |
|
| Name(s) found: |
sptl-2 /
CE27380
[WormBase]
|
| Description(s) found:
Found 20 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 586 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
metabolic process
[IEA tetrapyrrole biosynthetic process [IEA biotin biosynthetic process [IEA biosynthetic process [IEA |
| Molecular Function: |
pyridoxal phosphate binding
[IEA transferase activity, transferring nitrogenous groups [IEA glycine C-acetyltransferase activity [IEA transferase activity [IEA 5-aminolevulinate synthase activity [IEA 8-amino-7-oxononanoate synthase activity [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTSEVAKRNS PFRFLHKLPI RLTFTEKKSP LLISDLAKQR EENDELKKRT HILECAWNQE 60
61 HDDDEEEEEV KVDQGSEETT SSHDIYGDVG KRAPNEFEKI DRLTTIKVYI AWLALFVFAH 120
121 IREYVTRFGF VKDLSSKENA KMKDFVPLFS DFEAFYQRNC YIKVRDVFER PICSVPGATV 180
181 DLVDRVSHDG NWTYEYPGTR TNVINVGSYN YLGFAQSAGP CAEQSASSID REGLSCCTTV 240
241 HERGRSVSQA KLEKLVAEFL GVEDAICFSM GFATNSMNAP CLVDKHSLII SDKYNHASLI 300
301 LGCRLSGAST KVFEHNDMES LERILRDAIA YGNPKTHRPY KKILIIVEGI YSMEGSICNL 360
361 PGIIALKKKY QAYLYLDEAH SIGAMGKTGK GVVEYWGCDP KDVDILMGTF TKSFGAAGGY 420
421 IAGSKRTVDH LRAASPTGYY SSPMSPPIAQ QIYTSMSIIM GKDGTKDGAQ RIERLARNSH 480
481 YFRMKLKQNG FIVYGSNDSP VVPMLIYFPT MCGFYGREML ARNIGCVVVS FPATHMTEGR 540
541 VRFCISAAHT KEQLDEVLET VNLLGSMSRS KFSKRSHLYK NQKIEW |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
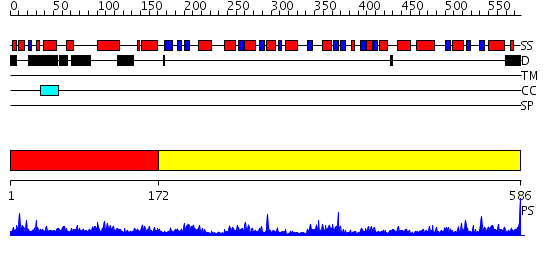
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..171] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [172..586] | 87.522879 | 2-amino-3-ketobutyrate CoA ligase |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.76 |
Source: Reynolds et al. (2008)