













| General Information: |
|
| Name(s) found: |
ddx-23 /
CE26887
[WormBase]
|
| Description(s) found:
Found 18 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 730 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA]
|
| Biological Process: |
DNA repair
[IEA embryonic development ending in birth or egg hatching [IMP DNA recombination [IEA |
| Molecular Function: |
nucleic acid binding
[IEA ATP binding [IEA ATP-dependent helicase activity [IEA helicase activity [IEA ATP-dependent DNA helicase activity [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAAEPPSLEE LLEKKKREED ELAKPKFLSK AERAALALKR REEEVAKLRE TQKQAEEARK 60
61 NWGRTEKTSD DRRDRGLDRG NDRDRDRERE RDRDRRRDRR ERSGSRDRRR RSRSRDRDGG 120
121 RRRDRSRSRD RRDRNDDRKR DRNEDSDNEE DVSKMADAVK DRYLGKQKEK KKRGRRLHEK 180
181 KFVFDWDAGE DTSQDYNKLY QSRHEIQFFG RGSVAGTDVN AQKKEKNSFY QEMMENRRTV 240
241 DEKEQEMHRL EKELKKEKKV AHDDRHWRMK ELSEMSDRDW RIFREDFNIS IKGGRVPRPL 300
301 RNWEEAGFPD EVYQAVKEIG YLEPTPIQRQ AIPIGLQNRD VIGVAETGSG KTAAFLLPLL 360
361 VWITSLPKME RQEHRDLGPY AIIMAPTREL AQQIEEETNK FGKLLGIKTV SVIGGASRED 420
421 QGMKLRMGVE VVIATPGRLL DVLENRYLLL NQCTYVILDE ADRMLDMGFE PDVQKVLEYM 480
481 PDTNMKKDTD EFDNEEALMK GFSTREKYRQ TVMFTATMSS AIERLARQYL RRPAVVHIGS 540
541 AGKPTERVEQ VVYMVPEDRK RKKLVEVLES QFQPPIIIFV NQKKGADMLS KGLTKLGFKP 600
601 TVLHGGKGQD QREYALQALK EGTSDILVAT DVAGRGIDVK DVSLVLNYDM AKSIEDYTHR 660
661 IGRTGRAGKH GKAITFLTPD DTAVYFDLKQ VLVESPVSSC PPELANHPDA QSKPGVFTSK 720
721 KRQDETLFLK |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
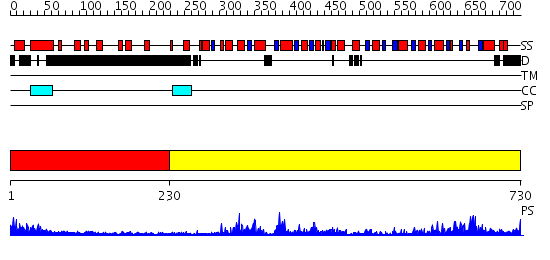
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..229] | 1.09 | BRCA2 helical domain; BRCA2 tower domain; OB-fold domains of BRCA2 | |
| 2 | View Details | [230..730] | 102.0 | No description for 2db3A was found. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)