













| General Information: |
|
| Name(s) found: |
sma-3 /
CE25974
[WormBase]
|
| Description(s) found:
Found 23 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 393 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA intracellular [IEA |
| Biological Process: |
positive regulation of cell size
[IMP nematode larval development [IMP positive regulation of organ growth [IMP tail tip morphogenesis [IMP regulation of transcription, DNA-dependent [IEA positive regulation of multicellular organism growth [IGI |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MNGLLHMHGP AVKKLLGWKI GEDEEKWCEK AVEALVKKLK KKNNGCGTLE DLECVLANPC 60
61 TNSRCITIAK SLDGRLQVSH KKGLPHVIYC RVWRWPDISS PHELRSIDTC SYPYESSSKT 120
121 MYICINPYHY QRLSRPQGLN SSMPSPQPIS SPNTIWQSSG SSTASCASSP SPSVFSEDGG 180
181 EVQVHQRPPP FRHPKSWAQI TYFELNSRVG EVFKLVNLSI TVDGYTNPSN SNTRICLGQL 240
241 TNVNRNGTIE NTRMHIGKGI QLDNKEDQMH IMITNNSDMP VFVQSKNTNL MMNMPLVKVC 300
301 RIPPHSQLCV FEFNLFFQML EQSCNDSDGL NELSKHCFIR ISFVKGWGED YPRQDVTSTP 360
361 CWLELRLNVP LAYIDQKMKQ TPRTNLMEPN SMT |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
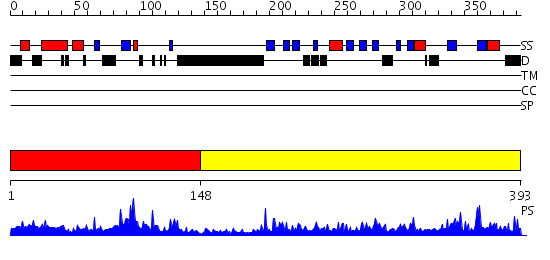
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..147] | 55.39794 | Crystal structure of Smad3-MH1 bound to DNA at 2.4 A resolution | |
| 2 | View Details | [148..393] | 91.522879 | Smad1 |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)