













| General Information: |
|
| Name(s) found: |
CE24787
[WormBase]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Caenorhabditis elegans |
| Length: | 752 amino acids |
Gene Ontology: |
|
| Cellular Component: |
intracellular
[IEA |
| Biological Process: |
nematode larval development
[IMP positive regulation of growth rate [IMP post-embryonic body morphogenesis [IMP embryonic development ending in birth or egg hatching [IMP regulation of transcription, DNA-dependent [IEA RNA processing [IEA tRNA 5'-leader removal [IEA growth [IMP |
| Molecular Function: |
transcription factor activity
[IEA ribonuclease P activity [IEA sequence-specific DNA binding [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MFIEVEKFVE ARTNSIAQLL KAIDNDALVS GEVTKGPRTA AQRLPRHMRR RAMAYDIRRF 60
61 PRTMREFAAA HLISKHAKKC PSRFARRKSA NSRTKFGRST STKGIWLSTH VWHAKRFRMI 120
121 QKWGFKLADR SFQRGFRAVL RDSNKNCVIR DRSYYTCVTI QCTDAYSKIS QFSQKGCRGQ 180
181 SSRDEEEECH QLYMPGKYPF GYIGPARFQR LSGDKVHIWI HPSSKLQFMK ALLEYYNLEK 240
241 QENTEEDLYK NSEIKVSIDA ENICRFHLSG PKCLSKLRDS IRFVNDEKFS SEHNYFLSVS 300
301 DSVFQSSRDG EVINLLIEDP RTTWDRKTVL KHKKVNKEAD NERIRVSKFW NKEFREMAIE 360
361 KRMADSEFHK QNSSKINGVI STEAKVPIIL IIRNEGLKAL DGADILIPEP FAKDFWVSLQ 420
421 RRGVRASGLR DEYAAHLESK ALYYPLDDVG SEAGRESELA MKKELIEKYL GKPHNRRCKH 480
481 WSAVSVKYPF EFKWDELSQD WNLSNKPRSE AFVCRDLQKL RIIEEAMKKG SGLEEFQEPG 540
541 MLIPVKLQFF GRGRPKKYGM VCLPTDDDLI SIRKNRNREI IQTPPEASSQ DSDIVEAMEI 600
601 EEITVKKNQG FMSLEAAASE KPINLKLLFE ETTKLDDKTT KRKRVNRKKR ESKKRRKIEQ 660
661 EKRKEEAEVE EVQKLATKYR FSANREIIGR LVAGEQSVLA GHGVGIGYIC ANTLSLIASN 720
721 YHKSKTVVMV RNSTSKYYHP AYVTILKNAT KI |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
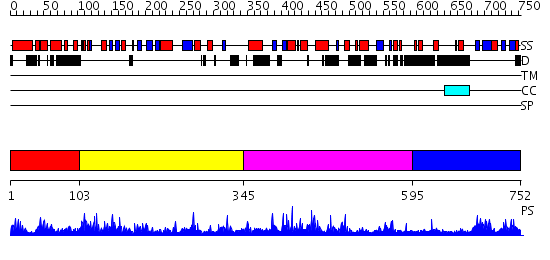
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..102] | 32.79588 | No description for PF06978.2 was found. No confident structure predictions are available. | |
| 2 | View Details | [103..344] | 36.69897 | Crystal Structure of a Component of Glycine Cleavage System: T-protein from Pyrococcus horikoshii OT3 at 1.5 A Resolution | |
| 3 | View Details | [345..594] | 2.1 | Crystal Structure of a Component of Glycine Cleavage System: T-protein from Pyrococcus horikoshii OT3 at 1.5 A Resolution | |
| 4 | View Details | [595..752] | 1.032973 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||
| 1 | No functions predicted. | ||||||
| 2 |
|
||||||
| 3 |
|
||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.80 |
Source: Reynolds et al. (2008)