













| General Information: |
|
| Name(s) found: |
gsy-1 /
CE24302
[WormBase]
|
| Description(s) found:
Found 19 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 672 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
glycogen biosynthetic process
[IEA locomotory behavior [IMP nematode larval development [IMP embryonic development ending in birth or egg hatching [IMP post-embryonic body morphogenesis [IMP |
| Molecular Function: |
glycogen (starch) synthase activity
[IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MPDHARMPRN LSSNKIAKTI AGEDLDEEEV LEMDAGQSAR EEGRFVFECA WEVANKVGGI 60
61 YTVLRSKAQI STEELGDQYC MFGPMKDGKW RLEVDPIEPE NRTIRAAMKR FQADGFRCMY 120
121 GRWLIEGYPK VILFDLGSGA VKMNEWKHEL FEQCKIGIPH EDIESNDAVI LGFMVALFLK 180
181 HFRESVTSYT PLVVAHFHEW QAGVGLLMTR LWKLDIATVY TTHATLLGRH LCAGGADLYN 240
241 NLDSFDLDAE AGKRKIYHQY CLERAACQTA HIFTTVSEIT GLEAEHFLCR KPDVLTPNGL 300
301 NVVKFAALHE FQNLHAQNKE KINQFIRGHF HGHLDFDLDK TLYFFTAGRY EFSNKGGDMF 360
361 IESLARLNHY LKTTSDPRHM GVTVVAFLIY PAPANSFNVE SLKGQAVTKQ LKEAVDRIKE 420
421 KVGQRIFDIC LQGHLPEPEE LMSPADNILL KRCIMSLHNS SLPPICTHNM IRADDPVLES 480
481 LRRTSLFNKP EDRVKVVFHP EFLSSVSPLI GLDYEDFVRG CHLGVFPSYY EPWGYTPAEC 540
541 TVMGIPSVST NLSGFGCFMQ EHVEDHEQKG IYVIDRRHKA AEESVQELAQ VMYDFCGQSR 600
601 RQRIILRNSN EGLSALLDWQ NLGVFYRDCR RLALERLHPD VDKIMRDNEG KVPSAATSRR 660
661 PSIHSSDGED DE |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
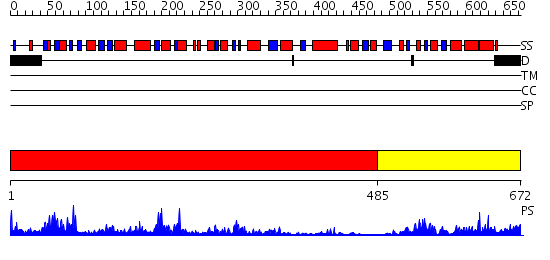
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..484] | 54.39794 | Crystal structure of the glycogen synthase from A. tumefaciens in complex with ADP | |
| 2 | View Details | [485..672] | 16.69897 | No description for 2gejA was found. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)