













| General Information: |
|
| Name(s) found: |
CE20567
[WormBase]
|
| Description(s) found:
Found 18 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 689 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
negative regulation of vulval development
[IMP nematode larval development [IMP embryonic development ending in birth or egg hatching [IMP reproduction [IMP |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MGSVITKETV LQENIYLKKL SGLEPVDDYD PFWNKLLSFS LKFDDDDEDT RIALESALDD 60
61 HLQCLMYNTQ TTGNFAAFIR LFLRRATELK TSEQCENKIY LWQTSNALLI LRYIARFLTQ 120
121 RMTEKEFVRI FAKSHESTET ESSSTSSSSD EEDDDEQKSE NENNNGATKY IPIVFQNTAE 180
181 EFTYELVSIL INISVNETTL AIHVEAVRCL LTLLSSQLYN ESIVNTSIVF RFFIDGSCAK 240
241 HAASLTKTLL LNYLIHNSEY HMTVQKPQES IVFGLASSMW SMVQMATGID SAEEEKKPPL 300
301 TLGNLSILLL LNLSCHQPLN SSNPFKETLA LFQNAQEVST LPTQIVSFKI DYNGLYERLC 360
361 ATAGQEPPML LLYMLLQANS GFRNYVLSRI NLENLVVPVL RILHDGVATS NNSHHVYLAL 420
421 IVALILSEDD IFCKIIHETM IKDLGWLDSD FSVREISLGG LTTLVLVRAI QKNALKTRDR 480
481 YLHTNCLAAL ANMSAFFKNL APIVCQRLIS LLDLLTKRHA KMVDHMRVSS QNDVADGQPI 540
541 NFHDDITALE EGIRTLLEII NSALCGGLRH NSHLIYNLLY HRALFDAYVQ HPMFQDLLVN 600
601 IAAVISHFSS KVIHVPAGDG STMLQIIEKE ANIWPTDRLA KFPELKFRYV EDEYTVDFFV 660
661 PYVWRLSVQH SGIYFETSRI KIFNAQSIA |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
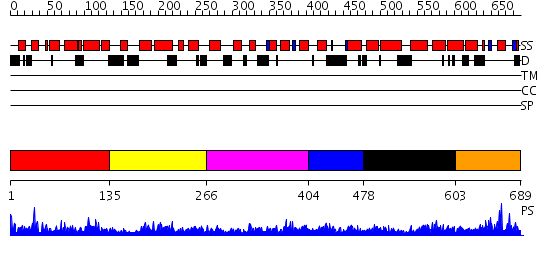
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..134] | 1.05097 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [135..265] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [266..403] | 2.057977 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [404..477] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [478..602] | 3.05698 | View MSA. No confident structure predictions are available. | |
| 6 | View Details | [603..689] | 1.029983 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.94 |
Source: Reynolds et al. (2008)