













| General Information: |
|
| Name(s) found: |
daf-5 /
CE18985
[WormBase]
|
| Description(s) found:
Found 20 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 627 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IEA |
| Biological Process: |
negative regulation of transcription from RNA polymerase II promoter
[IGI |
| Molecular Function: |
protein binding
[IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSDSPIGSSQ QVEPEHRTPD LMDIDPLIAN LKALHEETRS DDDDDGQPST SAKRKDSRAD 60
61 GIVIHQKKYS DPGRFLWIWL LGVRVPALSI DGEPHLPIEI LDDMLTKKDK KDQMSFQNLL 120
121 RYKNVYIRMA SPSQFRAVME KSKECENLNI TSLSLMSRSD IERIMGELRL ESMLTLAEHD 180
181 NWDISDRVHV VHVNFIDYCS EWLESDDLEE DVMQSGTHGY WYKNRRNMRC IECQHCEGKF 240
241 TPTDFIMHHH YPIKPSGFVH TGCNSFQWIR LIEVFDKSNE NLEAWNKFVL NSHRAGKREY 300
301 DEAAPHQAPP KRPAMETPVP VAADNGWEAD EEEEGEEIVD RDADIEKCKL RNKKKMENLH 360
361 IADFLGPSGS KGLKPRNKFE AVIIEQLNKM DDAALEALFL KSPEEYNLWV KESDFTHKVV 420
421 TQQQEWKAKM KDPNFKSRAS ANFDVSKGEF DNMRHFDNAS KATRQEIQQL AEQFANLDRD 480
481 AKLLTPMEFV LREHALLKNV SADAIRVLCN RPPLPPLPPP PPPPKPKPAP VQPISLGNIN 540
541 FVALAQQLIA SGIKLPLPIV TPPVVSTPAP VITPIPAALP ISPNSDFLKQ QLSTAMSSPA 600
601 LLSLYPKLTA GAYEQLAQFI KTTTVKN |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
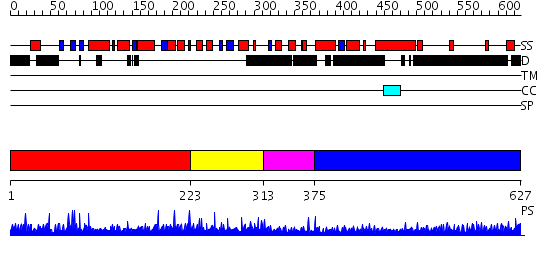
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..222] | 1.0 | Retinal determination protein Dachshund | |
| 2 | View Details | [223..312] | 1.93 | SMAD4-binding domain of oncoprotein Ski | |
| 3 | View Details | [313..374] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [375..627] | 32.213997 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.94 |
Source: Reynolds et al. (2008)