













| General Information: |
|
| Name(s) found: |
atg-7 /
CE18887
[WormBase]
|
| Description(s) found:
Found 21 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 647 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
dauer larval development
[IGI autophagy [IGI Mo-molybdopterin cofactor biosynthetic process [IEA meiosis [IMP |
| Molecular Function: |
ATPase activity
[IEA ATP binding [IEA catalytic activity [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MATFVPFVTC LDTGFWNEVN KKKLNDWKLD ETPKCISSQL SLHQTEGFKC HLSLSYDSLS 60
61 SLESTTGLSM SGTLLLYNTI ESFKMVDKSD LIRSEAEKIW ESITTRKWLQ NPRLLSQFFI 120
121 IAFADLKKFK YYYWTCVPAL VYPSEIKQEI TPLSSLGADH KILFDFYRKN NFPIFLYSKQ 180
181 SSKMLELSEL ENNTNPDEIC VVVADPSPVA YSAGWMVRNV LAAVAHLHPT WKHCHIISLR 240
241 SADSIGIKYT WTLPSAECSA DGAQNAVPKA VGWERNANDK LQPISVDLSK EFDPKILMER 300
301 SVDLNLSLIK WRLHPDIQLE RYSQLKVLIL GAGTLGCNIA RCLIGWGVRH ISFLDNSTVS 360
361 YNNPVRQSLS EFEDARLGRG KAETAQAAIQ RIFPSIQATA HRLTVPMPGH SIDEKDVPEL 420
421 EKDIAKLEQL VKDHDVVFLA LDSREARWLP TVLASRHKKI AISVAIGFDT YVIIRHGIGS 480
481 RSESVSDVSS SDSVPYSQLS CYFCSDVTAP GNSTFDRTLD QQCTVARPGT SMIASGIAVE 540
541 LLSSVLQYPD PLKTPASHDD NTTVLGAAPH QIRGFLGRFQ QILPSVKRFD QCVACGDAIA 600
601 AQFQQNGWKF VRDVMNSPGR LEEVTGLDEL QNSVNAIDID FEDDEDF |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
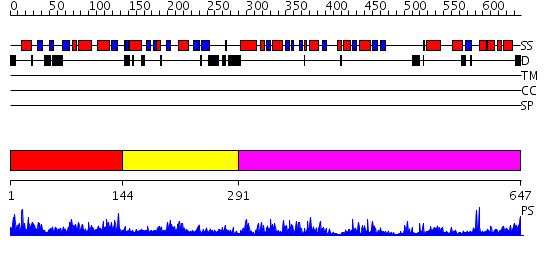
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..143] | 1.044974 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [144..290] | 2.207995 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [291..647] | 62.30103 | Molybdenum cofactor biosynthesis protein MoeB |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||
| 3 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.86 |
Source: Reynolds et al. (2008)