













| General Information: |
|
| Name(s) found: |
prmt-3 /
CE18281
[WormBase]
|
| Description(s) found:
Found 18 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 680 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IEA |
| Biological Process: |
rRNA modification
[IEA |
| Molecular Function: |
rRNA methyltransferase activity
[IEA rRNA (adenine-N6,N6-)-dimethyltransferase activity [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAEPAQETPC FSMEKAERAF LDGNFEEALK HFFVCIRMLP VERRGVYEDQ FAAALHGWIA 60
61 LDHNNASRAV CLYPQIKELF PETIRTKIAL IRAVQGTEQS KWLLNCLPIC KDSMDLATKP 120
121 EDLVALRIAR VNLTTIPFPQ WHIRMINDVK RNEAFAKALN DTIKSRITVV FDIGSGTGIL 180
181 SAIAARKTNL VTALEENMCL TMISKEVLKR NGVESRVNVH AKNSTYFETC EKADIVVSET 240
241 LDCCVFGEKI VETFLDAHVR FSHDRTIFIP HQATVYVRLF SCREIFDIHC QDYGGVRYRS 300
301 EYVKIGESDA EQPYWCASAA DYSQFELLSG SVAMHSVDFS SIANLKDSLA NSNSFKIRPA 360
361 KDGVAHGFSV HFTSDLTGHG DNIIDSSKSR AWDLGIIPFK EPCLVKCDKE YEGSWKLLNN 420
421 RLDVYNDFYD ENLQKHEDSL RYETASLDQL QKIRDDSYFK AMMGEIDPIE LPFTRDISSS 480
481 IPAQCVLNTV ESQTPIRHLI TNFCRYDGTL DQETFFRIEE FVGFRDHVQK IVPTKFRILG 540
541 HLFYSDRVHF DARPDPSAHC QVDLSTVRSF NLREMRDIRL TQREDIVMAS KEFVLYDFNI 600
601 SAEKFRQDTY ITVTQDVTVQ PTRPIADGVI YEFEILGIRN TKLRPVAAFL FPERVNTNPD 660
661 MTVVFDLHLG EMLISLKDLP |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
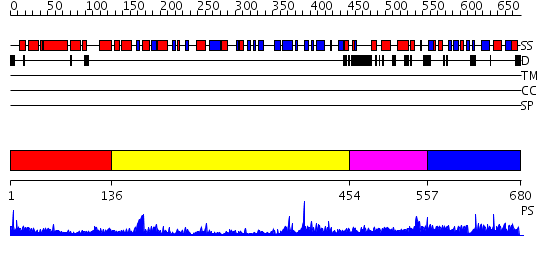
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..135] | 14.69897 | Crystal Structure of RumA, the iron-sulfur cluster containing E. coli 23S Ribosomal RNA 5-Methyluridine Methyltransferase | |
| 2 | View Details | [136..453] | 69.221849 | Structure of the Predominant protein arginine methyltransferase PRMT1 | |
| 3 | View Details | [454..556] | 1.073987 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [557..680] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||
| 1 | No functions predicted. | |||||||||||||||
| 2 |
|
|||||||||||||||
| 3 | No functions predicted. | |||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)