













| General Information: |
|
| Name(s) found: |
CE14894
[WormBase]
|
| Description(s) found:
Found 18 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 793 amino acids |
Gene Ontology: |
|
| Cellular Component: |
small-subunit processome
[IEA |
| Biological Process: |
positive regulation of growth rate
[IMP embryonic development ending in birth or egg hatching [IMP gamete generation [IMP rRNA processing [IEA |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTEPIKEIAA KRSVEAVYTG GTVRWSANGQ KLFSTCTNVV KVIDLADNTA SYTIGDPEDE 60
61 LRITCTAVDK NRNRLLVAYN NQIIREYSIP LDPSTTKPEL ARTWKTIHTA PILVMEFNEE 120
121 GHLLATGSAD HIVKVWNLTE QQCTHTLKGP SVVSAILFGK HEKLVVGYIE GQLNLYNIMR 180
181 GAPKKLVNTW KSHNSHITAL LQVPDSRVVV ALSRDQTMSI HETETQETLR VLPLYESIEG 240
241 GAIGHNGNLF TVGEEGVVKE FVIETAKLVR KKRIASSNLD HLAYDVVSNR FLAISAEQNL 300
301 IILDFENLKI ARQIVGFHDE IYSCCLLGKD ESHMAVASNT SEIRLYNTKT LDCQLIRGHT 360
361 ESVLSVVSPT WDTSLLASCS KDNSIIFWRL VTSPENDSCS TLVPVAAATG HANTVTALAI 420
421 SNTGRAPFLA SVSTDCTIKL WGLGVSTKLK EVKIDAEKDF VEQLPKLTCS STMVAHGKDV 480
481 NCVDISESDA LIATGGMDKL VKLWQVDTHK MQLGIAGTLS GHRRGVGDVK FAKNSHKLAS 540
541 CSGDMTIKIW NISEKSCLQT ISGHSCAVFR VIFARNDSQL ISADSAGIIK IWTIKTADCE 600
601 CSIDAHTDKI WSLSKNHNES EFVTAGTDGR IVVWKDVTEE KQKAEDNKRR EAIEQDQTLT 660
661 NLLSQKRYSE ALVFALTLAK PFCAFKVINA LMAEETPEEL GSAIRRLDTR QIEILLQFTV 720
721 KWNTNSRTSS VAQRVLYEIV HIVDPEELVS MPGAYGYIES FLPYTQRHMD RLDRAKQDVS 780
781 LFDFVWRQMR SSV |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
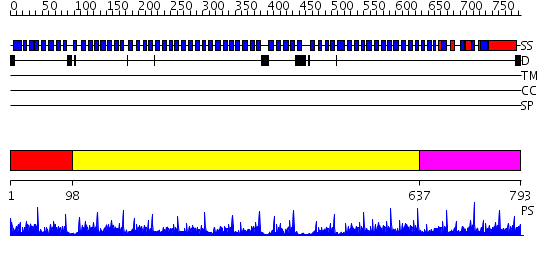
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..97] | 57.0 | Tup1, C-terminal domain | |
| 2 | View Details | [98..636] | 61.221849 | Actin interacting protein 1 | |
| 3 | View Details | [637..793] | 74.39794 | No description for 2gnqA was found. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)