













| General Information: |
|
| Name(s) found: |
dnj-22 /
CE14038
[WormBase]
|
| Description(s) found:
Found 18 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 296 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
response to heat
[IEA locomotory behavior [IMP nematode larval development [IMP morphogenesis of an epithelium [IMP positive regulation of growth rate [IMP embryonic development ending in birth or egg hatching [IMP gamete generation [IMP protein folding [IEA positive regulation of embryonic development [IMP growth [IMP |
| Molecular Function: |
heat shock protein binding
[IEA ATP binding [IEA unfolded protein binding [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MPLKPGANPY KILHLEKGCT DKEIQKAYRA QCLKWHPDKN LDNKEEAERR FIEAKEAFDF 60
61 LYDKEKREEY DNREERIRMA QEHHSKRMAE ADGKRKKLIE DLEKREKEFS NGGKRSADGS 120
121 TPMTAAQQAK KKKTDQRNFK EEIEAIRRQL EKEVNEEVKQ KATLMKTERE KHQKSQEKLT 180
181 PRLLLKWKTS EGLDYSEDDI RKLFSTFGII SHISSAIEKK DRRKRILEFE SGANAWGAEL 240
241 ETGNAPMPEI TCEWLQPPVE TVKKASDEPV KSTNTGANNL EHMSLDDLEA QIMGGF |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
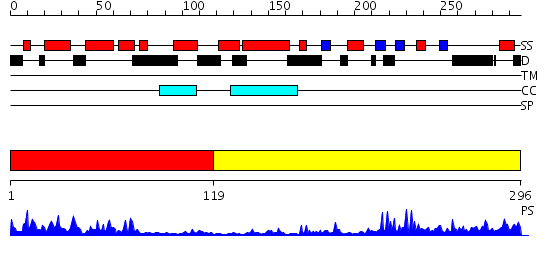
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..118] | 24.69897 | J-DOMAIN (RESIDUES 1-77) OF THE ESCHERICHIA COLI N-TERMINAL FRAGMENT (RESIDUES 1-104) OF THE MOLECULAR CHAPERONE DNAJ, NMR, 20 STRUCTURES | |
| 2 | View Details | [119..296] | 21.221849 | The crystal structure of Hsp40 Ydj1 |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)