













| General Information: |
|
| Name(s) found: |
gad-1 /
CE13295
[WormBase]
|
| Description(s) found:
Found 19 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 620 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
establishment of mitotic spindle orientation
[IMP cell migration involved in gastrulation [IMP embryonic development ending in birth or egg hatching [IMP regulation of mitotic cell cycle [IMP |
| Molecular Function: |
protein binding
[ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDDNTAPKKN SEKKARVFDL QAMFAQTISN APRTEEPTHV PQSVSVSSKS ESGVSSSAVS 60
61 DDEDDFMPDL PPGFQMQQSE AGPSSSRAEG PEDSDDSDFD ETEAISIIKL IPAACEAKIS 120
121 HGTQAISALR VEPPGVRFAS GGLDYYVKLF DFQKMDMSMR YDKELLPAES HVINSLAFSP 180
181 NGETLVVASG EAIIRLLDRA GKQWSETVRG DQYIIDLNIT KGHTATVNCV EFNPLNKNEF 240
241 ISCSDDGSLR LWNLDDHKVI TKCINKHRKV IKTKGANGKR VSPQVCTFSP DGKWIAAGCD 300
301 DGSVQAWKYG SQYVNVNYLV RKAHNGSITS IAFSPDSKRL LSRGFDDTLK MWSLDNPKEP 360
361 LLVKTGLENA FKSTDCGFSP RAEVVFTGTS SPNKDTPGTL QFFDPMTFDL VYKIDYPGIS 420
421 CHRIQWHPRL NQIIAGLSDG TIHVYYDQTM SQRGVMSCVT KPLKRNRASE VVREDMVLSP 480
481 LSLEMFQPRG EEGEEKEVTG WRIKKYLRMQ DNKLRPEFRK PADMPINGKS ANGRVAASGG 540
541 SLHSYLAKQI GTARNAEFLK DTDVRASILK HAKDAEENPL YIDKAYRKTQ PKKIFQETAV 600
601 EPEDQDDEEL QPVFKMPRTK |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
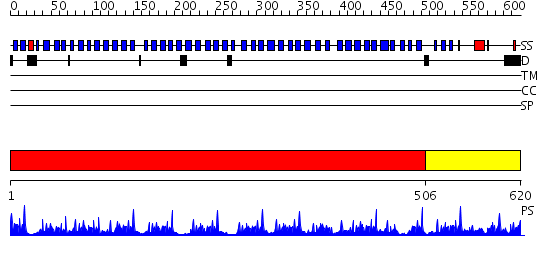
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..505] | 56.0 | Actin interacting protein 1 | |
| 2 | View Details | [506..620] | 22.69897 | No description for 2pm9A was found. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)