













| General Information: |
|
| Name(s) found: |
fum-1 /
CE11580
[WormBase]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Caenorhabditis elegans |
| Length: | 501 amino acids |
Gene Ontology: |
|
| Cellular Component: |
tricarboxylic acid cycle enzyme complex
[IEA |
| Biological Process: |
purine ribonucleotide biosynthetic process
[IEA protocatechuate catabolic process [IEA positive regulation of growth rate [IMP embryonic development ending in birth or egg hatching [IMP arginine biosynthetic process via ornithine [IEA aspartate metabolic process [IEA fumarate metabolic process [IEA reproduction [IMP |
| Molecular Function: |
N6-(1,2-dicarboxyethyl)AMP AMP-lyase (fumarate-forming) activity
[IEA 3-carboxy-cis,cis-muconate cycloisomerase activity [IEA catalytic activity [IEA aspartate ammonia-lyase activity [IEA argininosuccinate lyase activity [IEA fumarate hydratase activity [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSAVSMLQGE MLARGGAVIA RGASLATARN FSRTTVPMAK IRKERDTFGE LEVPADKYYG 60
61 AQTARSQMNF KIGGPEERMP IPVIHAFGIL KKAAALVNTE FGLDKKLADA ISQAADEVVD 120
121 GKLDEHFPLV TWQTGSGTQS NMNVNEVISN RAIEILGGEL GSKKPVHPND HVNMSQSSND 180
181 TFPTAMHIAV GREVNSRLLP ALKKLRTALH NKAEEFKDII KIGRTHTQDA VPLTLGQEFS 240
241 AYVTQLDNSI ARVESTLPRL YQLAAGGTAV GTGLNTRKGF AEKVAATVSE LTGLPFVTAP 300
301 NKFEALAAHD ALVEVHGALN TVAVSFMKIG NDIRFLGSGP RCGLGELSLP ENEPGSSIMP 360
361 GKVNPTQCEA ITMVAAQVMG NQVAVSVGGS NGHFELNVFK PLIVRNVLQS TRLLADSAVS 420
421 FTDHCVDGIV ANKDNIAKIM RESLMLVTAL NPHIGYDNAA KIAKTAHKNG TTLVQEAVKL 480
481 GILTEEQFAQ WVKPENMLGP K |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
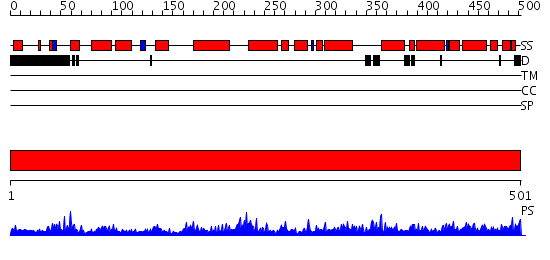
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..501] | 157.0 | RECOMBINANT YEAST FUMARASE |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.82 |
Source: Reynolds et al. (2008)