













| General Information: |
|
| Name(s) found: |
ugt-23 /
CE08282
[WormBase]
|
| Description(s) found:
Found 23 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 530 amino acids |
Gene Ontology: |
|
| Cellular Component: |
integral to membrane
[IEA |
| Biological Process: |
carbohydrate metabolic process
[IEA antibiotic metabolic process [IEA metabolic process [IEA lipid glycosylation [IEA |
| Molecular Function: |
transferase activity, transferring hexosyl groups
[IEA carbohydrate binding [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MQLLSALLLQ ILFNVSSPYK ILVYSNLFGH SHIKFVGSVV DTLVDAGHDV TVVLPVIDST 60
61 QKNRTSIKSG AKQIIIDADE EVVEIFSQTE KFLSNMWTME MSNPLMMWMN PRILTQLFGK 120
121 QCTKVMKLSE LLDQLKSEKF DLAITEAFDS CGYGIFEYLQ IPAHVSILSC ARMDHVSDVL 180
181 GQPIAPSYVP GTQSLYSDRM TMTQRFLNYL QFLNGNSMFS DIGDYESANA KKLLGVERSW 240
241 REILPESSFL LTNHIPVLEF PAPTFDKIVP IGGISVNKNK ETLKLEHYFD TMVSMRKKNV 300
301 IISFGSNIKS MDMPDEYKKS LAELFQLMPD VTFIWKYENL ADKKYTCGIM NINRVEWIPQ 360
361 TELLADSRVD AFITHGGLGS VTELATMGKP AVVIPIFADQ TRNAEMLKRH GGAEVLHKTD 420
421 LANPETLRKT LRKVMDDPSY RQNAQRLAEM LNNQPTNPKE TLVKHVEFAA RFGKLPSMDP 480
481 YGRHQNIIEY YNLDILAIAT ILLTLVVYLK IFAVGVVVRC CCGKKKVKFE |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
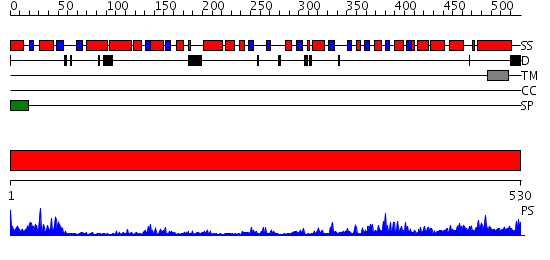
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..530] | 60.0 | No description for 2pq6A was found. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Source: Reynolds et al. 2008. Manuscript submitted |

|