













| General Information: |
|
| Name(s) found: |
erm-1 /
CE07805
[WormBase]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Caenorhabditis elegans |
| Length: | 563 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoskeleton
[IEA cytoplasm [IEA cell cortex [IDA filamentous actin [IDA membrane-enclosed lumen [IDA extrinsic to membrane [IEA |
| Biological Process: |
locomotory behavior
[IMP morphogenesis of an epithelium [IMP nematode larval development [IMP positive regulation of growth rate [IMP embryonic development ending in birth or egg hatching [IMP protein amino acid dephosphorylation [IEA actin filament organization [IMP gamete generation [IMP tube formation [IMP growth [IMP reproduction [IMP positive regulation of multicellular organism growth [IMP |
| Molecular Function: |
protein tyrosine phosphatase activity
[IEA structural molecule activity [IDA cytoskeletal protein binding [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSKKAINVRV TSMDSELEFA IQSSTTGKQL FDQVVKTIGL REIWYFGLQY TDNKGFPTWL 60
61 KLNKKVLSQD VKKDPTLVFK FRAKFYPEDV AEEIIQDVTM RLFYLQVKDG ILSDEIYCPP 120
121 ETSVLLASYA MQAKYGDYVP ETHVAGCLTA DRLLPQRVLG QFKLNSEEWE RRIMTWWADH 180
181 RATTREQAML EYLKIAQDLE MYGVNYFEIR NKKGTDLYLG VDALGLNIYD KADRLSPKVG 240
241 FPWSEIRNIS FNDKKFVIKP IDKKAHDFVF YAPRLRINKR ILALCMGNHE LYMRRRKPDT 300
301 IEVQQMKQQA REDRALKIAE QEKLTREMSA REEAEQRQRD AEKRMAQMQE DMERARLELA 360
361 EAHNTIHSLE AQLKQLQLAK QALEQKEYEL RELTAQLQSE KAMSDGERRH LRDQVDARER 420
421 EVFSMREEVE RQTTVTRQLQ TQIHSQQHTQ HYSNSHHVSN GHAHDETATD DEDNGATELT 480
481 NDADQNVPQH ELERVTAAEK NIQIKNKLDM LTRELDSVKD QNAVTDYDVL HMENKKAGRD 540
541 KYKTLRQIRG GNTKRRIDQY ENM |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
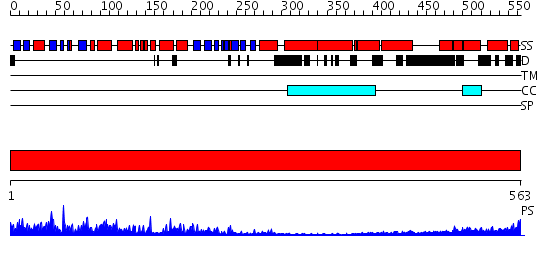
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..563] | 91.0 | No description for 2i1jA was found. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)