













| General Information: |
|
| Name(s) found: |
glh-3 /
CE07736
[WormBase]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Caenorhabditis elegans |
| Length: | 720 amino acids |
Gene Ontology: |
|
| Cellular Component: |
P granule
[IDA |
| Biological Process: | NONE FOUND |
| Molecular Function: |
nucleic acid binding
[IEA ATP binding [IEA zinc ion binding [IEA protein binding [IPI DEAD/H-box RNA helicase binding [IPI ATP-dependent helicase activity [IEA JUN kinase binding [IPI helicase activity [IEA protein self-association [IPI |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDKSPTKTSI RTKFARHQPI SDVDTTEQSS SCIKKDDRGL SSFGVQSSVF SRRSCRMSEL 60
61 EAKPTIISED QRIAVRSEIG GSFSGFDDKV DNVFHSNNNL HGSPSTTELE CPGIMNPRFL 120
121 VGRSLNSRSR AVTRGSKRTS NVKENEGSIH RSDDQVSTEN CSAKDEERDR DSGGVSSYGN 180
181 KRSDEFCGTS PILEAKGFGI SNTCFNCKKY GHRATECSAP QRECANCGDP NHRANECASW 240
241 SKNGVQEPTK VTYVPVVDKM EEVFSMLKIN AGDFFDKFFD ASVQLVSRGQ PVTIQPCKSF 300
301 SDSDIPQSMR RNVERAGYTR TTPIQQYTLP LVADGKDILA CAQTGSGKTA AFLLPIMSRL 360
361 ILEKDLNYGA EGGCYPRCII LTPTRELADQ IYNEGRKFSY QSVMEIKPVY GGINVGYNKS 420
421 QIMKGCTIIV GTIGRVKHFC EDGAIKLDKC RYLVLDEADR MIDSMGFGPE IEQIINYKNM 480
481 PKNDKRQTMM FSATFPSSVQ EAARKLLRED YTMITIDKIG AANKCVIQEF ELCDRTSKVD 540
541 KLLKLLGIDI DTYTTEKNSD VFVKKTIVFV AQQKMADTLA SIMSAAQVPA ITIHGAREQK 600
601 ERSAALKLFR SGAKPVLIAT AVVERGLDIK GVDHVINYDM PNNIDDYIHR IGRTGRVGNS 660
661 GRATSFISLA DDVQILPQLV RTLADAEQVV PSWMKEAAGG TSNPNKFEKS IDTEEPEEAW 720
721 |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
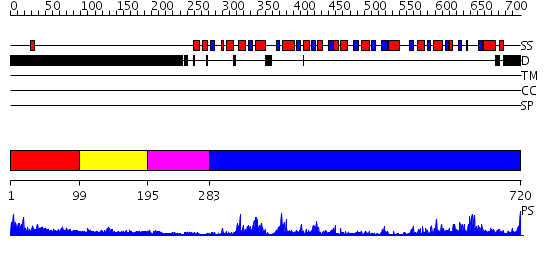
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..98] | 3.098989 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [99..194] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [195..282] | 2.28 | Solution Structure of the Zinc-finger domain in LIN-28 | |
| 4 | View Details | [283..720] | 104.0 | No description for 2i4iA was found. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)