













| General Information: |
|
| Name(s) found: |
CE07461
[WormBase]
|
| Description(s) found:
Found 16 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 357 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
biosynthetic process
[IEA |
| Molecular Function: |
pyridoxal phosphate binding
[IEA transferase activity, transferring nitrogenous groups [IEA catalytic activity [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MRLSFFDGIH VASPIKELHT SELFQKEICP VKINLAIEAY RTEDGEPWVL PVVREIELKF 60
61 PHEPHHNHEY LPILGHDGFC KSATALLLGN DSLAIKEGRS FSVQCISGTG AICVGAEFLA 120
121 QVLSMKTIYV SNPCCLCYNP TGMDPTREQW IQMAQVIKQK NLFTFFHIAD QGLASGDADA 180
181 DAWAVRFFVE QGLEMIVSQS FSKNFGLYND RVGSLTVIVN KPSHIANLKS QLTLVNVSNF 240
241 SNPPAYGARI VHEILKSPKY REQWQNSIKM MAFRIKKTRQ ELIRELNMLQ TSGKWDRITQ 300
301 QSGLFSYTGL TPCQVDHLIA HHKIYLLSDG RINICGLNMS NLDYVARAID DTVRTIH |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
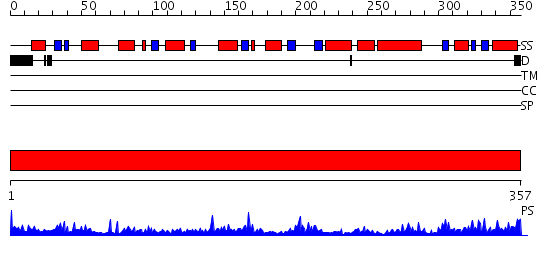
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..357] | 79.69897 | Aspartate aminotransferase, AAT |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.92 |
Source: Reynolds et al. (2008)