













| General Information: |
|
| Name(s) found: |
pccb-1 /
CE07269
[WormBase]
|
| Description(s) found:
Found 20 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 536 amino acids |
Gene Ontology: |
|
| Cellular Component: |
acetyl-CoA carboxylase complex
[IEA |
| Biological Process: |
sodium ion transport
[IEA fatty acid biosynthetic process [IEA |
| Molecular Function: |
acetyl-CoA carboxylase activity
[IEA methylmalonyl-CoA decarboxylase activity [IEA ligase activity [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MLSRFARQLA LPVFNGVLHP TRATSSIAHT IKVAHKIDET REKALLGGGK KRVDAQHARG 60
61 KLTARERIDL LLDRNTFREY DMFAEHTCND FGMQKEKYPG DSVVTGRGEI NGRTVFVFSQ 120
121 DFTVFGGSLS SIHAKKIVKI MREAMLVGAP VIGLNDSGGA RIQEGVESLA GYADIFQENV 180
181 MASGVVPQIS MIMGPCAGGA VYSPALTDFT FMVRDTSYLF ITGPDVVKAV TNEEVTQEEL 240
241 GGAKTHTVTS GVAHGAFDND VDALMSLREL FNYLPLSNTD AAPIRAAEDP WDRAVPSLDT 300
301 VVPLESTAAY NMKDVVHALV DEGDFFEIMP DYAKNLVIGF ARMNGRTVGI VGNNPKFAAG 360
361 CLDINSSVKG ARFVRFCDAF NIPIITLVDV PGFLPGTAQE YGGIIRHGAK LLYAFAEATV 420
421 PKITIITRKA FGGAYDVMSS KHLRGDINYA WPTAEVAVMG AKGAVSILFR NDKEHAVQHE 480
481 EEYTELFSNP FPAAVRGFVD DIIIPSETRK KVCEDLNMLE SKKLKNPWKK HGNIPL |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | hcp2 ip from december | Cheeseman, I.M., et al. (2005) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
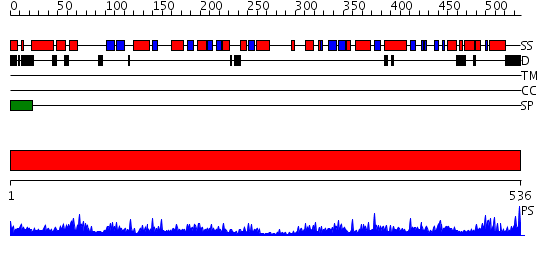
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..536] | 166.0 | Crystal structure of propionyl-CoA carboxylase, beta subunit (TM0716) from THERMOTOGA MARITIMA at 2.30 A resolution |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Source: Reynolds et al. 2008. Manuscript submitted |

|