













| General Information: |
|
| Name(s) found: |
nrs-1 /
CE05684
[WormBase]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Caenorhabditis elegans |
| Length: | 545 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IEA |
| Biological Process: |
tRNA aminoacylation for protein translation
[IEA nematode larval development [IMP embryonic development ending in birth or egg hatching [IMP asparaginyl-tRNA aminoacylation [IEA lysyl-tRNA aminoacylation [IEA aspartyl-tRNA aminoacylation [IEA reproduction [IMP growth [IMP translation [IEA |
| Molecular Function: |
nucleotide binding
[IEA nucleic acid binding [IEA ATP binding [IEA aminoacyl-tRNA ligase activity [IEA lysine-tRNA ligase activity [IEA asparagine-tRNA ligase activity [IEA aspartate-tRNA ligase activity [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSKLYIDTDA GSDQHPGTEA QPLATLVQAM LISKNSGEFL MKKKSEEGES WEPAAKAAIK 60
61 KAVKKYEAEV KKLEKAGCRE KEAEEAQHAA LEEAKKITFS LDKSLPEAKV IKIGESVQHR 120
121 DQRVSIKAWV HRLRRQGKSL MFLVLRDGYG FLQCVLNDKL CQSYDAVTLS TETSVQVYGI 180
181 IKALPDGKSA PDGHELTVDY WEVIGKAPAG GIDNVLNESA GVDVMLDNRH LVIRGENASR 240
241 ILRIRAAATR AMRDHFFAAG YTEVAPPTLV QTQVEGGSTL FGLDYYGEPA YLTQSSQLYL 300
301 ETCNAALGDV YCISQSYRAE KSRTRRHLSE YQHVEAECAF ITFDQLMDRI EALVCDTVDR 360
361 LLADPVTKSL IEFVNPGYKA PARPFKRMPY KEAIEWLQKN DVRNEMGEKF VYGEDIAEAA 420
421 ERRMTDTIGV PILLNRFPHG IKAFYMPRCA DDNELTESVD LLMPGVGEIV GGSMRIWKED 480
481 QLLAAFEKGG LDSKNYYWYM DQRKYGSVPH GGYGLGLERF ICWLTDTNHI RDVCLYPRFV 540
541 GRCAP |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
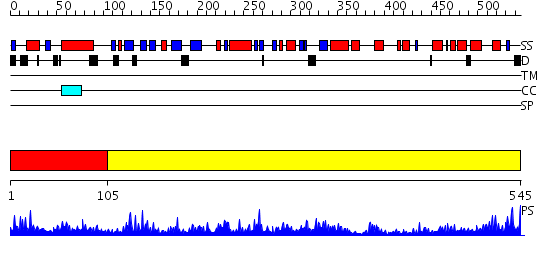
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..104] | 1.89 | Crystal structure of pectate lyase Pel9A | |
| 2 | View Details | [105..545] | 109.0 | No description for 1x54A was found. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)