













| General Information: |
|
| Name(s) found: |
CE05376
[WormBase]
|
| Description(s) found:
Found 19 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 674 amino acids |
Gene Ontology: |
|
| Cellular Component: |
proton-transporting two-sector ATPase complex
[IEA membrane [IEA |
| Biological Process: |
proton transport
[IEA |
| Molecular Function: |
nucleic acid binding
[IEA zinc ion binding [IEA protein binding [IEA hydrolase activity, acting on acid anhydrides, catalyzing transmembrane movement of substances [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSESAEVISN ADASYGTDTD TLPDTELPSL NVLEVDEPKE EIKKNEMSRI DIIREAADNV 60
61 QSCRVCYDEY HSSSNQARVL QCGHTFCTRC VVGCSSTMNN TSEEFGIKCP ECRKINEQVP 120
121 ATVPINFQLM QILTTLSLVK VLQTPPQDQE IPKYENFERL GTEIPIADLV NLTMGELYEH 180
181 LKAVFNAIRL RGKRDKTSIR PSVVHRYDSE FKKIDEVERS MNRVLHNVIK LQSGETLPRP 240
241 APTITIPYWQ LDREYRRSLQ DNDWMREMVV FRPPTSGPVP VRPLVAPPQP PAQPPVQDVD 300
301 TVLEREQAAR AEADPRRHLN AVREAHLRRE RESAQEREAA VRNEQLMRQL LDRNENMQNV 360
361 HIDRNDPFSL FRHLQRQEER DLEMHLRFRR TLDQDRDRES MRRVADGLPP ILDHDDVDVE 420
421 EDDDESVASD NSSFVDEPVE RPPPPRMTLG EAVTIYRMIE VFSPRHFRHG PDSFSLPHNH 480
481 ANLNSLYREL LPPNSPITIT DTMSDSQLAD LRRYSQANNV DCDRLLRVLR HHSIKFAPTI 540
541 VESPRLTERR RRRQRPVPAD DYNAHLSVIR KRAKEALRGN RPEQPTNGEE SNLTIRLIVN 600
601 LANSMGVREK LDDVICPQHR AQEEEKEMFV PRTNGSTRQE MLFCNVCRCE VPMGSKAVHS 660
661 QGKRHIAAMA KFVF |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
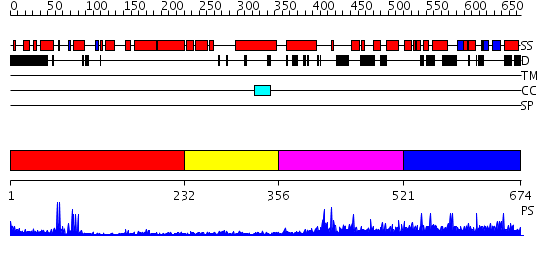
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..231] | 9.522879 | RAG1 DIMERIZATION DOMAIN | |
| 2 | View Details | [232..355] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [356..520] | 28.522879 | No description for 2iwgB was found. | |
| 4 | View Details | [521..674] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||
| 1 |
|
||||||
| 2 | No functions predicted. | ||||||
| 3 | No functions predicted. | ||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)