













| General Information: |
|
| Name(s) found: |
coh-1 /
CE04726
[WormBase]
|
| Description(s) found:
Found 19 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 652 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA nuclear chromosome [IEA |
| Biological Process: |
locomotory behavior
[IMP embryonic development ending in birth or egg hatching [IMP synaptonemal complex assembly [TAS chromosome segregation [IMP meiosis [IMP |
| Molecular Function: |
damaged DNA binding
[ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MCHPIEFCIC TISDASAHTS SSRTSFCTPS TSHIVSPSFP ICIAAKCSRC GCVAAMFYAD 60
61 FVLSKKGPLS KVWLAAHWEK KLSKAQIFET DVDEAVNEIM QPSQKLALRT TGHLLLGICR 120
121 VYSRKTKYLL ADCNEAFLKI KLVFRSGALD QPNPVLPTFS IQDIYGDFGD NVLPEFDEEE 180
181 LNHAPICQSR LDDITLKEDI PQKFTYNYGE PLEDDDFGEI AAGVGPEDYY RLMEDVNKMD 240
241 LEMELARDAA TTSDNLFGRE REPTPFLNDA HAPGNAIFGD EDFAGGDMDD DDHNVQGSSG 300
301 FTDNNAMHME DMDYESGNTM MRGETPLRSD TPNTFRAPSP APSIAPSVAT SVSAMEHDTI 360
361 ESYYERQAQK RAMQKEQNMR KRRVDDVKMI TGEEMKGNMA DFSDILTRLD LAPPSRKLMM 420
421 GKKRSHAEYM HHNPGMIGFS KNKQFIRKYQ SYLVVTKHDD VDEKNEWIKN ALGLREIGEE 480
481 ELQQQQQEMD MHVQDDSSYV DDFDEVPPLD FDQLDMMDMP QSMDDINMVD DDVPQRNPLS 540
541 PFAPMVEDEE MSPSEKRKKR DEEKEEPETD EDNRWSKRTH ALLQNIATKL ENQNGQVELD 600
601 EMLKKGTSRK VAAAKFYSLL CLKKNQCIDI EQKEPYGDIM IKAGPNININ TI |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
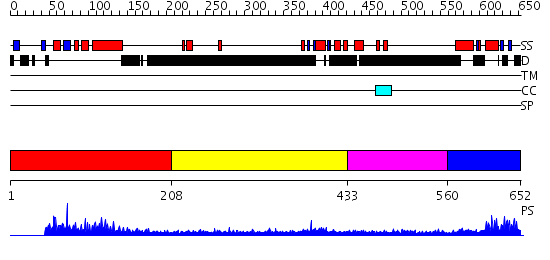
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..207] | 60.568636 | No description for PF04825.4 was found. No confident structure predictions are available. | |
| 2 | View Details | [208..432] | 1.161993 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [433..559] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [560..652] | 9.0 | Sc Smc1hd:Scc1-C complex, ATPgS |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||
| 4 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.83 |
Source: Reynolds et al. (2008)