













| General Information: |
|
| Name(s) found: |
CE04010
[WormBase]
|
| Description(s) found:
Found 20 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 422 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
arginine metabolic process
[IEA ectoine biosynthetic process [IEA tetrapyrrole biosynthetic process [IEA biosynthetic process [IEA biotin biosynthetic process [IEA gamma-aminobutyric acid metabolic process [IEA |
| Molecular Function: |
pyridoxal phosphate binding
[IEA 4-aminobutyrate transaminase activity [IEA transaminase activity [IEA glutamate-1-semialdehyde 2,1-aminomutase activity [IEA diaminobutyrate-pyruvate transaminase activity [IEA adenosylmethionine-8-amino-7-oxononanoate transaminase activity [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MLSSLRRVVP SLPRGSRSLT SQQIFDREKK FGCHNYKPLP VALSKGEGCF VWDVEGKKYF 60
61 DFLAAYSAVN QGHCHPKLLK VVQEQASTLT LTSRAFYNNV LGEYEEYVTK LFKYDKVLPM 120
121 NTGVEACESA VKLARRWAYD VKGVKDNEAV VVFAENNFWG RSIAAISAST DPDSFARFGP 180
181 FVPGFKTVPY NNLKAVEDAI KDKNVAAFMV EPIQGEAGVV LPDPGYLKGV SDLCKKYNVL 240
241 FITDEVQSGL GRSGKLLAHY HDNVRPDIVV LGKALSGGFY PVSAVLCDDN VMMNIKPGEH 300
301 GSTYGGNPLA CKVAIAALEI LQEEKLVENS AVMGDLLMSK LKTLPKDIVS TVRGKGLFCA 360
361 IVINKKYDAW KVCLKLKENG LLAKNTHGDI IRFAPPLCIN KEQVEQAADI IIKTVTDFAK 420
421 QN |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | hcp2 ip from december | Cheeseman, I.M., et al. (2005) |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
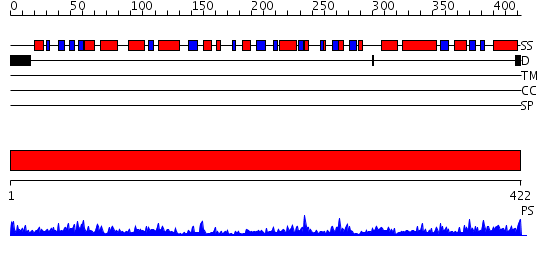
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..422] | 104.0 | Ornithine aminotransferase |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.96 |
Source: Reynolds et al. (2008)