













| General Information: |
|
| Name(s) found: |
nth-1 /
CE03559
[WormBase]
|
| Description(s) found:
Found 19 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 259 amino acids |
Gene Ontology: |
|
| Cellular Component: |
intracellular
[IEA |
| Biological Process: |
DNA repair
[IEA base-excision repair [IEA |
| Molecular Function: |
DNA binding
[IEA endonuclease activity [IEA DNA-(apurinic or apyrimidinic site) lyase activity [IEA DNA N-glycosylase activity [IEA 4 iron, 4 sulfur cluster binding [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MRKDMIAPVD TMGCHKLADP LAAPPVHRFQ VLVALMLSSQ TRDEVNAAAM KRLKDHGLSI 60
61 GKILEFKVPD LETILCPVGF YKRKAVYLQK TAKILKDDFS GDIPDSLDGL CALPGVGPKM 120
121 ANLVMQIAWG ECVGIAVDTH VHRISNRLGW IKTSTPEKTQ KALEILLPKS EWQPINHLLV 180
181 GFGQMQCQPV RPKCGTCLCR FTCPSSTAKN VKSETEETST SIEVKQEVED EFEDEKPAKK 240
241 IKKTRKTRTK IEVKTESET |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
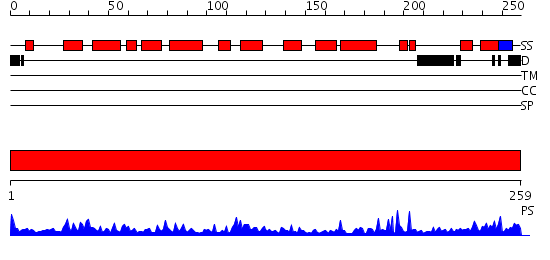
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..259] | 59.69897 | REFINEMENT OF THE NATIVE STRUCTURE OF ENDONUCLEASE III TO A RESOLUTION OF 1.85 ANGSTROM |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)