













| General Information: |
|
| Name(s) found: |
din-1 /
CE03148
[WormBase]
|
| Description(s) found:
Found 18 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 567 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IEA proteinaceous extracellular matrix [IEA |
| Biological Process: |
nematode larval development
[IMP positive regulation of growth rate [IMP embryonic development ending in birth or egg hatching [IMP post-embryonic body morphogenesis [IMP cell adhesion [IEA blood coagulation [IEA regulation of transcription [IEA |
| Molecular Function: |
nucleic acid binding
[IEA binding [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSAEEAATVM AVASSDPNPP ATSTVDLAAM LQQLQAAQAA QAAQQVPVVT TASTPNPLSN 60
61 LETLLSTASL ANLATGGALN PLSMLALTSS LNQSSPVYQG IARVLLTMNM GQMLATHQTS 120
121 ELLATMNQQE TLMALLAARN GLPFAMPQQN QQPQMPAQGG FAIPTVLPHM SLKRNAKDQL 180
181 SVGGVSDRKK SCPLHAMIGQ GQQPPPPQQP MQAVAPAPPR SPSPPRKSMF ENLPPEMKEK 240
241 NEMFRKEILR RLDIILLEEL GAEDEEDQKP DLKQIPTSEE DTDDSKADSM GAEGSAFRRI 300
301 LSRSSTMGNN SGSPSASGTT SPSTSSSISS GPDSPPLEGE PLNSEFMDML TEVAQKHREQ 360
361 SNTDALSAKI VDEQSFSQHF PMVWTGRLAL KSTEAMINLH LINGSETFLN DVLGRQVTEE 420
421 NPRRDSVKIL QRLRLDNGQV EHIYRILTNP EYACCLALSS VNNIENLKEN DTNLKSHFID 480
481 YLINKKIAGI SSLGEVETKF KSARVHVFAP GEIVNRYLSE LATSLHDYLQ NTDTRYLLIV 540
541 FTNDKADPNM TGPPSVASLA VPPVSST |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
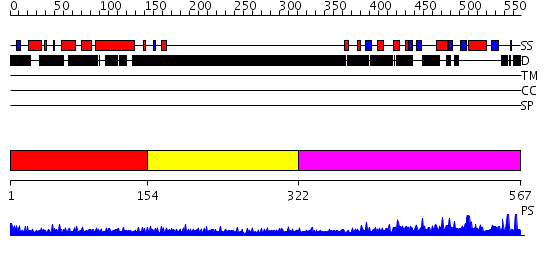
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..153] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [154..321] | 6.213996 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [322..567] | 50.30103 | Crystal structure of the SPOC domain of the human transcriptional corepressor, SHARP. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.64 |
Source: Reynolds et al. (2008)