













| General Information: |
|
| Name(s) found: |
CE01108
[WormBase]
|
| Description(s) found:
Found 20 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 784 amino acids |
Gene Ontology: |
|
| Cellular Component: |
intracellular
[IEA |
| Biological Process: |
nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
[IEA |
| Molecular Function: |
nucleic acid binding
[IEA 3'-5' exonuclease activity [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MGDTTSSEDV PENKQKSLKF EIIDARMKIF KDIVKSKSSE SVKEEQIYQK SLEFFDEDLK 60
61 SSEESVSNEE IKTGSEKELE PLNVFDIILI MLQQLPERKK PIGSLLSKWI LMNFMNWMQD 120
121 KQSIMEQQMT EYYQKKAGLA CVKEPKNEEY LLQIFKISKE FVIDLRKSKV QEYLENQKFK 180
181 EAAEIVMKHE VVDDYSFEQI TLPLILCDKV QIVDELLKIS KKLQKSYISF LDQFVAETDE 240
241 TVNAFFEPYK EKGMVTINLS RFHGKSLTIF MQKFFNGQVK QFKFDLEERR DAPKFVANMK 300
301 RKALKYFVGK RFEDHEMNDE LFCEHMKSTL PECTDKTIVQ FLILLWDTCI YERRIEALFW 360
361 ATYSNIDRNS KYMPPDLKEE LENPTTEMKN GLEKLQALRT TKNCQEEDEQ LYVFEEQKKY 420
421 PIRIVQNEQD LEILLSELGE LEEGMYIGYD SEFKPYHLID VSTSRLAIIQ LFFKDKAWLI 480
481 NCVAIDNLAS RDDVWIRLYK GLFESNKFSI VGFDIRQDIE AMFTVPSINK NFKIENIQNV 540
541 ICVKSLAENV NALSMDILNL STKTSKLSVL ADHLVGLKMD KSEQCGNWQC RPLRRNQIIY 600
601 AVMDAVAVFE VFQKIVEVVR KHELDAEKLL VESHMITVKK EKVRRDCKNI SLIPWNEFYQ 660
661 IIHTHRNPEK PLQKPSELKI VVDTMVLGLG KNLRLLGFDV YIPRDVTELK EFLRKMDKME 720
721 ESEQRLVISV PSRSYEMLKS DNPNAKFVLI PNIYEKVPID LVCSFFDFFN IDISPDQDYI 780
781 KLNC |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
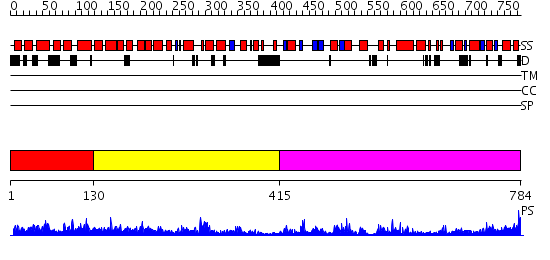
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..129] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [130..414] | 27.0 | STRUCTURE OF TAQ DNA POLYMERASE | |
| 3 | View Details | [415..784] | 68.0 | STRUCTURE OF LARGE FRAGMENT OF ESCHERICHIA COLI DNA POLYMERASE I COMPLEXED WITH D/TMP |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||
| 1 | No functions predicted. | |||||||||||||||
| 2 |
|
|||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)