













| General Information: |
|
| Name(s) found: |
tag-165 /
CE00868
[WormBase]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Caenorhabditis elegans |
| Length: | 682 amino acids |
Gene Ontology: |
|
| Cellular Component: |
thylakoid membrane
[IEA |
| Biological Process: |
electron transport
[IEA |
| Molecular Function: |
NADP or NADPH binding
[IEA FMN binding [IEA oxidoreductase activity [IEA FAD binding [IEA ferredoxin-NADP+ reductase activity [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTDFLIAFGS QTGQAETIAK SLKEKAELIG LTPRLHALDE NEKKFNLNEE KLCAIVVSST 60
61 GDGDAPDNCA RFVRRINRNS LENEYLKNLD YVLLGLGDSN YSSYQTIPRK IDKQLTALGA 120
121 NRLFDRAEAD DQVGLELEVE PWIEKFFATL ASRFDISADK MNAITESSNL KLNQVKTEEE 180
181 KKALLQKRIE DEESDDEGRG RVIGIDMLIP EHYDYPEISL LKGSQTLSND ENLRVPIAPQ 240
241 PFIVSSVSNR KLPEDTKLEW QNLCKMPGVV TKPFEVLVVS AEFVTDPFSK KIKTKRMITV 300
301 DFGDHAAELQ YEPGDAIYFC VPNPALEVNF ILKRCGVLDI ADQQCELSIN PKTEKINAQI 360
361 PGHVHKITTL RHMFTTCLDI RRAPGRPLIR VLAESTSDPN EKRRLLELCS AQGMKDFTDF 420
421 VRTPGLSLAD MLFAFPNVKP PVDRLIELLP RLIPRPYSMS SYENRKARLI YSEMEFPATD 480
481 GRRHSRKGLA TDWLNSLRIG DKVQVLGKEP ARFRLPPLGM TKNSAGKLPL LMVGPGTGVS 540
541 VFLSFLHFLR KLKQDSPSDF VDVPRVLFFG CRDSSVDAIY MSELEMFVSE GILTDLIICE 600
601 SEQKGERVQD GLRKYLDKVL PFLTASTESK IFICGDAKGM SKDVWQCFSD IVASDQGIPD 660
661 LEAKKKLMDL KKSDQYIEDV WG |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
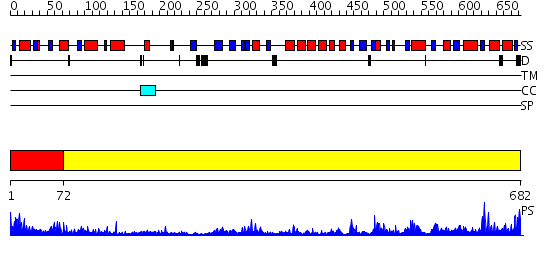
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..71] | 3.0 | A CRYSTALLOGRAPHIC STRUCTURAL STUDY OF THE OXIDATION STATES OF DESULFOVIBRIO VULGARIS FLAVODOXIN | |
| 2 | View Details | [72..682] | 111.0 | NADPH-cytochrome p450 reductase; NADPH-cytochrome p450 reductase, N-terminal domain |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.37 |
Source: Reynolds et al. (2008)