













| General Information: |
|
| Name(s) found: |
CE00231
[WormBase]
|
| Description(s) found:
Found 21 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 418 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
N-acetylglucosamine metabolic process
[IEA |
| Molecular Function: |
N-acetylglucosamine-6-phosphate deacetylase activity
[IEA hydrolase activity [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MLPRKLKIDS TELAEITNEL VQFLNCLVLR SGGLKKEHIW VRNGRILDER TVFFEEKTMA 60
61 DVQIDCEGLI LSPGFIDLQL NGGFGIDFST YNSDDKEYQE GLALVAKQLL AHGVTSFSPT 120
121 VITSSPETYH KILPLLKPSN ASSEGAGNLG AHLEGPFISA DKRGCHPEQL VITSLSPNPV 180
181 EIIEHVYGST ENIAIVTMAP ELEGAQEAIE YFVSTGTTVS VGHSSAKLGP GEMAVLSGAK 240
241 MITHLFNAMQ SYHHRDPGLI GLLTSSKLTP DHPLYYGIIS DGIHTHDSAL RIAYHTNSAG 300
301 LVLVTDAIAA LGMSDGVHKL GTQTIHVKGL EAKLDGTNTT AGSVASMPYC IRHLMKATGC 360
361 PIEFALQSAT HKPATLLGVS DEKGTLDVGR LADFVLIDKN VTVKATFCSG KRVFLAQD |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
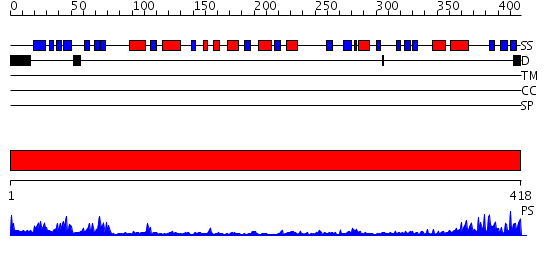
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..418] | 52.522879 | Crystal Structure of the N-Acetylglucosamine-6-phosphate deacetylase from Escherichia coli K12 |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)