













| General Information: |
|
| Name(s) found: |
CE00136
[WormBase]
|
| Description(s) found:
Found 21 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 608 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
sodium ion transport
[IEA nematode larval development [IMP positive regulation of growth rate [IMP embryonic development ending in birth or egg hatching [IMP RNA processing [IEA growth [IMP |
| Molecular Function: |
RNA binding
[IEA ribonuclease III activity [IEA methylmalonyl-CoA decarboxylase activity [IEA ligase activity [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MFRHVAQNLG SRNTSIQSYR LLRTRWERGY LKDLYHRRQI LGADPAISRS SYPNCSVQVR 60
61 NHHCSADKST LQWAPIKTSI DNSSDDFAAN TAEMKVLVED LKAKISKIEQ AGGEKAVKLH 120
121 RSRGKMLARE RIDGIVDAGS PFIEFSQLAG YEMYGKEEVP SGGILTGVGI VSGRVCVIVA 180
181 NDATVKGGTY YPITVKKHLR AQEIARENKL PCIYLVDSGG ANLPRQADIF ADSQHFGRIF 240
241 YNQATMSSEG IPQLAVVMGS CTAGGAYVPA MSDQAIIVKG TGTVFLGGPP LVKAATGEEI 300
301 SAEELGGADL HCGESGVTDY YAHNDKHALY LARSCIAGLP PVEEHMTFNP NADEPLYPAE 360
361 EIYGIVGSNL KKTYDVREVI ARIVDGSRFH EFKERYGETL VTGFATIYGQ RVGILANNGV 420
421 LFAESAMKGS HFIELCCQRK IPLLFLQNIT GFMVGRDAEA GGIAKHGAKL VTAVACAKVP 480
481 KITVLVGGSY GAGNYGMCGR GYSPRYVFMW PNSRISVMGG EQAANVLSTV QKEKKKREGA 540
541 DWTDQQDLEL RKPVEEKFEK EGHPYFASAR LWDDGVIDPK DTRKVLGLAF QSTLQKPIPE 600
601 TKFGVFRM |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
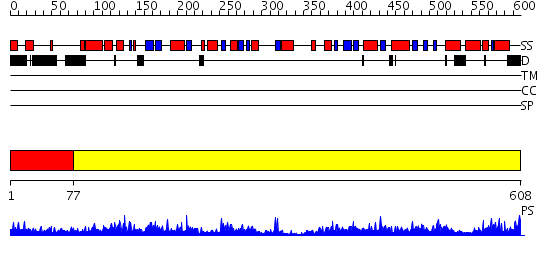
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..76] | 3.39794 | No description for 2d5dA was found. | |
| 2 | View Details | [77..608] | 149.0 | Crystal structure of propionyl-CoA carboxylase, beta subunit (TM0716) from THERMOTOGA MARITIMA at 2.30 A resolution |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)