













| General Information: |
|
| Name(s) found: |
cog4 /
SPCC338.13
[Sanger Pombe]
|
| Description(s) found:
Found 19 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 738 amino acids |
Gene Ontology: |
|
| Cellular Component: |
Golgi transport complex
[ISS nucleus [IDA cell division site [IDA cell tip [IDA |
| Biological Process: |
intra-Golgi vesicle-mediated transport
[ISS intracellular protein transport [IC retrograde transport, vesicle recycling within Golgi [ISS |
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDISINDCTD ISQIKQRFHD LQVESQRTDE KLERLLSDAQ PTEKFNSLIK NMAERLVLFV 60
61 GQIEELKDAF CNTTIVSEEV IERIKSVDRE QNRIKECLLF VRQVRDFKEC LQDLNRAMHH 120
121 QQWEKAADLV HRASSTSPAI IEGKFAHAVV PTAEQPLAPM DTLKEITESL HTLFWREFHK 180
181 AARNQDQKEI TRYFKLFPLI GKEKEGLEAY WHFFGGIIAS KARATLDEPP THALFFAQAF 240
241 TGLVEHVASI IRAHTPLVQK YYKAKNTITV IEKLQGDCDR QGSIIVNTMF DVRRIDNLVS 300
301 SIASYKYILL HAKLKNRAFV SDQKELERVS LQTLHPILNE MSAIVSKWNI SKIFISRLVL 360
361 RLSQSDGTSN DPSVQDNLIC ASIFNSSKME LLLKKQLLPS LLQLETYYFR RSIETSLELE 420
421 EYYTKVSPWM SSIVDDVMYV TKQVFQRAFF TVSSLFFTRF VNESLIPILR NDYYVYLSHN 480
481 LLTVCNIIKA QFQRLKNANS IPAKQVENYI TLVNSASLSK QYLKSIVDGV SSRLEEVFAF 540
541 AKDQKLVKKS IDNFLQLTVN FENLCKTSFN MYFPIFLLPR IEQCIDDSFD GINYVLSYED 600
601 YTKETEHERL VVTRLRSVWD RVLLLEQFTP ENQLSLRSMA CEKAASYIEN LILYKIQWND 660
661 YGAMALENDI SSLITIFSND QANLRHSFER LQEILILLVW ESDSTAPEQL INDLNLQLLS 720
721 IDIVSAIMEK KANVQGED |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
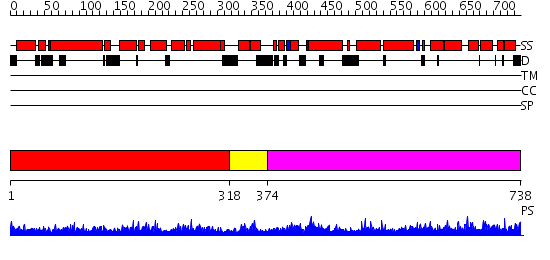
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..317] | 48.0 | Structure of unphosphorylated STAT5a | |
| 2 | View Details | [318..373] | 1.070995 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [374..738] | 1.5 | No description for 2fji1 was found. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)