













| General Information: |
|
| Name(s) found: |
mug72 /
SPCC1902.02
[Sanger Pombe]
|
| Description(s) found:
Found 20 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 574 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cell division site
[IDA cell tip [IDA cytoplasm [IDA |
| Biological Process: |
biological_process
[ND]
|
| Molecular Function: |
NADP or NADPH binding
[IEA]
oxidoreductase activity [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSEEDTSKLR ILSVGSNAIS AFISWRLSES KACHTTLIWR NRCESVLSEG IRIRSSVFGS 60
61 TKWKPDVVAP TVEQLAMNSE PFDYIFVCLK ILPSVYNLDT AIKEVVTPGH TCIVLNTTGI 120
121 VGAEKELQHA FPNNPVLSFV LPDQFAQRGP LQFEHTTFAA DSAKSVIYVG LTEEEDDVPD 180
181 SVQDAMIETL TLTLEAGGVS CDFLSKIQKK QWETGVGHMC FYPLSIINDE PNLALMYRLK 240
241 SFAKVIDGLM DEAFSIAQAQ GCEFEPEKLD VLKRHIVNRM LATPRPSYPY QDYIAHRPLE 300
301 VAVLLGYPVE IAKELGVSVP RMETLLALFD AKNKRNLTVR AGTPQSSPNF NPAMRRSPVG 360
361 AASRSPSRST IGISNRIGSV DDLLNTRQFT SSPIGGSMPK GPNSIYKIPS ASMVNLSSPL 420
421 VTSPSGLNPT GRPSRFGGRV RGNPLTMNKA GSVSDLLSTS TNMASSDALE TASMIGVPSA 480
481 MPPNSFDMLT LTQRRNRRNN QSSSPPAPSD RRYTMGARRP VQTRGMTDSV IPILEDPMST 540
541 LYDTSRYPTR NSKPATPKAS RPPSIASTVH RRMD |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
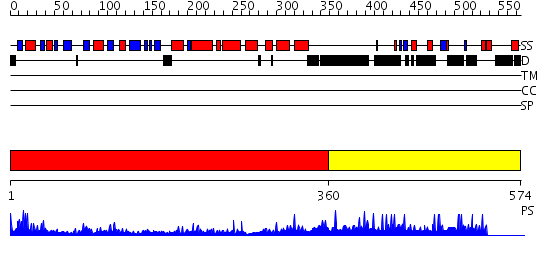
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..359] | 51.39794 | Crystal Structure of the Putative 2-Dehydropantoate 2-Reductase from Enterococcus faecalis | |
| 2 | View Details | [360..574] | 1.046919 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.90 |
Source: Reynolds et al. (2008)