













| General Information: |
|
| Name(s) found: |
SPBC13E7.03c
[Sanger Pombe]
|
| Description(s) found:
Found 18 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 713 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytosol
[IDA |
| Biological Process: |
nuclear-transcribed mRNA poly(A) tail shortening
[ISS]
DNA replication, Okazaki fragment processing [ISS] |
| Molecular Function: |
RNA binding
[ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MEVVEGRRKT CLDVVDSPSS RQKQTIAVSL VDGKNYPTRF LNSRTIDKLK QELNTSNLST 60
61 GEMSTNETLK NAQEVAGKLL SEENEDSISE HFDEYLGADK PGISFLTRLK TLESWFQSLS 120
121 LQDRLTTLRT LLHHLPSQEI STLLSSSLTS SPSNSGLSLD KSLPSSPKGD SPSLSSSLPS 180
181 LTTKSNLSGN LNMVTPASTQ GPAFSSKHGF SNALSTASPI PVRTSSVSST YLTQDREASS 240
241 KNCLSKALAF SSIEPPASSA STSPRNTPTP SNNGTSINAN VTSSLTSNST GKTSKTTDLL 300
301 IAASKKSLPS NSTPSKPNTS FFETPHNNIW DSRDRGAFSA PPAPFFPLGF SPHLNDESSR 360
361 SRWSNISYSP PPPPPPPELL NHSPKSRPLG DKPYLFYRNN QIGPRTRTEG RSSITEGKPF 420
421 LSSSLRFEHV PSANDLNSVV NARVEKSTGQ PSTPLNRQHY FNELPHSTTP VTLPSLIHGS 480
481 EIDRRRSGFC LNNFNSTPPF QPYHYEIGSG LPQQMHATNT ILTNPIDPNS NISTPVGMHL 540
541 CTLPYTYQPF SSVEKIETPP NNSKNQTYRR SSRGSNKTRK SISHSKNTDK HVGNELPQDI 600
601 PSWLRSLRLH KYTNNLKDTD WDALVSLSDL DLQNRGIMAL GARRKLLKSF QEVAPLVSSK 660
661 KMNENLKAAK NQSSESLTSF KGHTDSEDLP SGSMSNEISS NSTKQDVSSS SMD |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
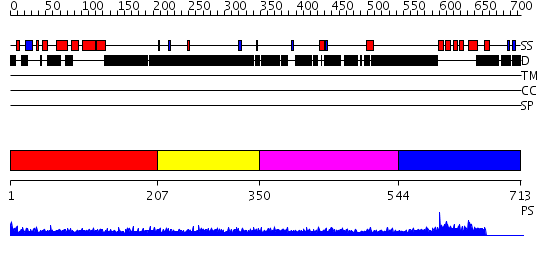
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..206] | 1.191995 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [207..349] | 2.197989 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [350..543] | 3.197997 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [544..713] | 30.0 | RNA recognition by the Vts1 SAM domain |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)