













| General Information: |
|
| Name(s) found: |
his7 /
SPBC29A3.02c
[Sanger Pombe]
|
| Description(s) found:
Found 18 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 417 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA cytosol [IDA |
| Biological Process: |
histidine biosynthetic process
[IGI |
| Molecular Function: |
ATP binding
[IEA]
phosphoribosyl-ATP diphosphatase activity [ISS phosphoribosyl-AMP cyclohydrolase activity [IGI |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MALLPFFDLT NFESDASEEL GWLKYVGRVQ TRVFPQHFKD NLEKVRKISE TIDVIVDTTA 60
61 ELGPEACVNL LNAGALAILV NEEMLNELAD ISPNRLVLKT DTTDIGKIEK LSQVAGSIQW 120
121 IGSAENYPPD FFERASKIIH KAVMPEGGGR TLYLEFPEQP SMEVLKSFSV HSVVPVLSSS 180
181 FLTVKPAEEP KKLSLADLIL ISANTDREDG LFSTLVVNEL GIALGLVYSS KESVAESLKT 240
241 GTGVYQSRKR GLWYKGASSG AVQHLIHIDV DCDEDCLRFV VYQTGKGFCH LDTLHCFGQA 300
301 SGLCQLEKTL IDRKNNAPEG SYTARLFSDP KLLRAKIMEE AEELCDATTK ENVIWEMADL 360
361 MYFAITRCVG SGVSLNDISR HLDLKHRKVT RRKGDAKVAW QEKLKDKGGV ANTSYTA |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
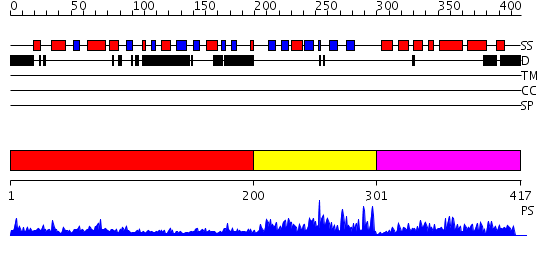
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..199] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [200..300] | 44.522879 | Crystal structure of Methanobacterium thermoautotrophicum phosphoribosyl-AMP cyclohydrolase HisI | |
| 3 | View Details | [301..417] | 22.0 | Crystal structure of Phosphoribosyl-ATP pyrophosphohydrolase from Bacillus cereus. NESGC target BcR13. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||
| 2 |
|
||||||||||||||||||
| 3 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)