













| General Information: |
|
| Name(s) found: |
wsp1 /
SPAC4F10.15c
[Sanger Pombe]
|
| Description(s) found:
Found 18 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 574 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cell tip
[TAS actomyosin contractile ring [TAS actin cortical patch [ISS |
| Biological Process: |
fungal-type cell wall biogenesis
[IMP contractile ring contraction involved in cell cycle cytokinesis [IMP cytokinesis, actomyosin contractile ring assembly [IMP actin nucleation [IDA establishment or maintenance of cell polarity [IGI actin filament polymerization [IMP actin cortical patch assembly [IGI |
| Molecular Function: |
protein binding
[IPI |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MPPSSSITQE DKATIRKYIP KSTNKIIAAA VVKLYVAYPD PNKWNYTGLC GALVLSYDTT 60
61 AKCCWFKLVD VVNNSGIIWD QELYQNMDYR QDRTFFHSFE LDKCLAGFSF ANETDAQKFY 120
121 KKVLDKGCHP ESIENPVLSF ITRKGSSRHA PNNSNIQPPS AAPPVPGKEN YNAVGSKSPN 180
181 EPELLNSLDP SLIDSLMKMG ISQDQIAENA DFVKAYLNES AGTPTSTSAP PIPPSIPSSR 240
241 PPERVPSLSA PAPPPIPPPS NGTVSSPPNS PPRPIAPVSM NPAINSTSKP PLPPPSSRVS 300
301 AAALAANKKR PPPPPPPSRR NRGKPPIGNG SSNSSLPPPP PPPRSNAAGS IPLPPQGRSA 360
361 PPPPPPRSAP STGRQPPPLS SSRAVSNPPA PPPAIPGRSA PALPPLGNAS RTSTPPVPTP 420
421 PSLPPSAPPS LPPSAPPSLP MGAPAAPPLP PSAPIAPPLP AGMPAAPPLP PAAPAPPPAP 480
481 APAPAAPVAS IAELPQQDGR ANLMASIRAS GGMDLLKSRK VSASPSVAST KTSNPPVEAP 540
541 PSNNLMDALA SALNQRKTKV AQSDEEDEDD DEWD |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
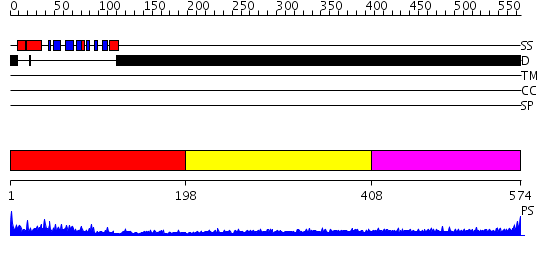
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..197] | 31.0 | No description for 2ifsA was found. | |
| 2 | View Details | [198..407] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [408..574] | 3.197997 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)