













| General Information: |
|
| Name(s) found: |
swi1 /
SPBC216.06c
[Sanger Pombe]
|
| Description(s) found:
Found 19 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 971 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA spindle pole body [IDA replication fork protection complex [TAS |
| Biological Process: |
DNA replication checkpoint
[IGI replication fork protection [IDA mating type determination [IMP replication fork arrest [IDA negative regulation of mitotic recombination [IDA |
| Molecular Function: |
protein binding
[IPI protein heterodimerization activity [IPI |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MELDEVIQGI VSAIGGFDYS DDEKVYVLGD EALACLKDLK RYLQVVDEKY KVWQIRSLLS 60
61 SLQLVTNDIC PILSDWDKDI TNYRNWRIAL ACVELLVPLT WPLETEHETF RENVDVLYNL 120
121 RQAQSNYKNS ILSYKKGSVL SAILAVLLKP LSTPAESRTL RDKGIIRIVL LLFRNILQID 180
181 ELKTKNETII SFAKAHILDL IVTLVSNLDE FEHFDVYILE IVYNLIRGCK PSALFSDASL 240
241 TNSQTELNSL LLKESTQNRY LKRNAHTRHN RFGTMLSVQT EDRRFTIASQ NIKTDGLDEL 300
301 DSHKRFRKRG TRRKHFDDIN KSFFINTEAG TALRNFAVEF LEAGFNPLFQ SLLKDLERED 360
361 PRVLPIHKMQ LLYVQSFFLE FMRFSSKPKK TEEIYSNDYS FGLAASVFDQ RALIMHNRLM 420
421 VESFEMKQWS TFQASMLSMT QLLFTLRSMT LCSSEIYQRI ADNLLSNIFY QEEILLLVYS 480
481 ALKHFKTQSF GYLDAITELT IVLLKELEKF SSAKQYLYVK KRRRNQKSVD SNVLESDEDE 540
541 ESSLINANAA VEDRLFDFGR YESRYCDNGC IDSFVLFLQC YQDLDSKQIH RAISFFYRIF 600
601 VKQKCHVYLY RLDFLRVLDK MFNDHVYFST TNSARQDFEQ FFVYYMRKLS DALKDVPALF 660
661 IELPFPKLTD TFYYLEYGKS PLFSIHGSRK GPLYETVPGL SHLEKVAAVV ACLINENKSD 720
721 LLDELKVQLN CLISERKLIT LADENKYINE GGNDGERMGK NLKGDTDSFN TALLKDGKFR 780
781 LLLELCGFEE SDNNIDVQAL WKLPNSVIID ELVEHAMLLR RFTDDPPTFE GTKPEDLLVR 840
841 KQRGNVRLPS SSEGETSDEE IEFEADDPIT FANRREALNK ITDRKRKKMK TNETIIDHTT 900
901 RKKKENHLRS AKYIVDSDDD SETDIAFFQS EAALREKNAQ KASALFKRID DLEMEGKLQE 960
961 IEQLSENSSS D |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
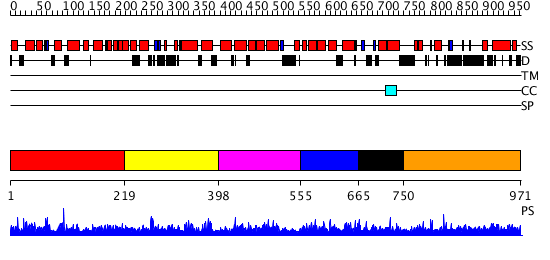
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..218] | 1.018976 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [219..397] | 1.02198 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [398..554] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [555..664] | 1.016971 | View MSA. Confident ab initio structure predictions are available. | |
| 5 | View Details | [665..749] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [750..971] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.94 |
Source: Reynolds et al. (2008)