













| General Information: |
|
| Name(s) found: |
sre1 /
SPBC19C2.09
[Sanger Pombe]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 900 amino acids |
Gene Ontology: |
|
| Cellular Component: |
SREBP-SCAP complex
[IDA nucleus [TAS integral to membrane [IEA] |
| Biological Process: |
negative regulation of gene expression
[IEP oxygen and reactive oxygen species metabolic process [IMP response to hypoxia [IMP positive regulation of ergosterol biosynthetic process [IEP positive regulation of heme biosynthetic process [IEP positive regulation of transcription via sterol regulatory element binding involved in ER-nuclear sterol response pathway [IMP positive regulation of lipid biosynthetic process [IEP regulation of transcription, start site selection [IEP |
| Molecular Function: |
DNA binding
[IDA transcription factor activity [IMP specific RNA polymerase II transcription factor activity [RCA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MQSSIPSVSV SVASPAMETP TKASPDSKSP NSVGAIPSSS PLASSTKAST STPFVENCSN 60
61 LLCDLASIVE DSPPTLTNTS LSPHSFSLSD MESSMSNWLN PFAFDNTMNS APPLFTSTNM 120
121 GSPNSLENST NPLLSNCGSP NSFQNETFTG PSLNEFDADD KIQRQMKILH SVDTIDPSTV 180
181 QNYPSADMSN PEVKLKTEEI ITPMDTTCKP EPSAKKIKLS PSSEDSCSIP ETLPFSAPKS 240
241 RGSLSPSETP DFVAGPGGKP KKTAHNMIEK RYRTNLNDRI CELRDAVPSL RAAAALRCGN 300
301 SLDDEDLGGL TPARKLNKGT ILAKATEYIR HLEAKNKELQ KTNKQLSDRL AFYEDPSMAP 360
361 PSNDTRAVNS VNVVSSSDYS VHQSSRPNLT QRAFTSPTLN TMGRTALNGM VGLGLFNYFG 420
421 NDSSQSVYGL FALPPFLMSP FTGTVLFNML KIGVVLLGLF YLLHDNSLFK GFKGEKKSKV 480
481 STRSSMSPSS ILFRKTVFEK YCLLDHSTST ISLFFGLLIF TLKSAYGYLT HRLSALYTSS 540
541 ENWVYSEQQL AEVRNMEKLL DAQLMGGDAK VDRLRLLMVF ASSFSLPPSS HTCALQAMYC 600
601 QLIFSNTSVP SAIVSKCVAF FWNAAKKQHS KSSVHAELRE LPECTANLIE NSHADDVFSP 660
661 NMVERLWVLA KCTRDSAQMS DSIISSLSDV LVLSPLEVLA SWYAADLLDA LLMESLSRKV 720
721 EISEIEEIIS LCPKNSSIIR HALLAKLVLF PENTADSLNE VLAAYKNTLD LCSQDKRKQS 780
781 SVLKINLSKL FTLHSCLSLA LQRLGYGDVS KRMYQEIFVP DSDADITPLS FIISWTALNT 840
841 FAPICTSPKE NDVVEKMAMY VRTAIGTLKI QDLKLSRKLI NSCIDIGSRL QEDLGYVSSA 900
901 |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
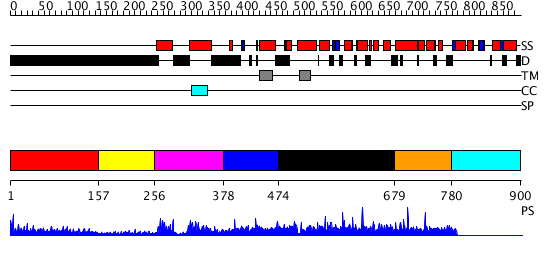
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..156] | 13.328997 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [157..255] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [256..377] | 21.154902 | Myc prot-oncogene protein | |
| 4 | View Details | [378..473] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [474..678] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [679..779] | N/A | Confident ab initio structure predictions are available. | |
| 7 | View Details | [780..900] | N/A | Confident ab initio structure predictions are available. |
| Source: Reynolds et al. 2008. Manuscript submitted |

|