













| General Information: |
|
| Name(s) found: |
mug71 /
SPBC577.12
[Sanger Pombe]
|
| Description(s) found:
Found 18 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 606 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA cytoplasm [ISS cytosol [IDA |
| Biological Process: |
biological_process
[ND]
|
| Molecular Function: |
ATP binding
[IEA]
endoribonuclease activity [ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MKVLGLISGG KDSCFNLMHC VSLGHEVVAL ANLHPEDGKD EIDSFMYQSV GHDVIPLYAE 60
61 CFDLPLYREK IGGQSINQNL DYQFTEKDET EDLYRLIKRV LTNHPDLEAV STGAILSTYQ 120
121 RTRVENVCKR LGLKSLSFLW QKDQEKLLND MVVSGLNAIL IKVAAIGLTR KDLGKSLAEM 180
181 QDKLLTLNKK FELHPCGEGG EYETLVLDCP LFKKRIVLTD KEVVEHSSGE VCYLKVKACV 240
241 KDKPEWQPIS LKSELVPNEE LLGEEYSHIY HTISKKYELI DDQEETPTSL IPIPLRESAF 300
301 QQKKGSFLVL GNVVATKGSY NTFQGEAESA INNLNELLGT YGYSNKNVYF VTVILSSMSK 360
361 FAEFNSVYNK YFDFTNPPSR SCVAAPLASE YRIVMSCIVG DVTEKRALHV QGQSYWAPAN 420
421 IGPYSQSICA NGVVFISGQI GLIPSVMELK LHDKIFEMVL ALQHANRVAK AMRVGSLIAC 480
481 LAYVCDSRDA DCVVKIWSEY TKNTGESSPV LVALVDALPR NASVEWQLLY NDSSCDVPLL 540
541 SSLVTNQTLF GSDTAWDVAL LNQNGLRMES SFIHHEHPSA YAIVLNNAFP NSQLLHVRYT 600
601 ARDQNV |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
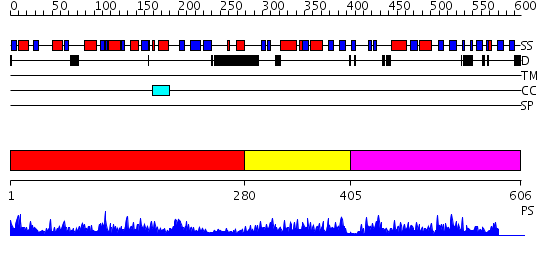
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..279] | 63.045757 | Crystal Structure of PH1257 from Pyrococcus horikoshii OT3 | |
| 2 | View Details | [280..404] | 37.522879 | Purine regulatory protein YabJ | |
| 3 | View Details | [405..606] | 44.39794 | No description for 2dyyA was found. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.91 |
Source: Reynolds et al. (2008)