













| General Information: |
|
| Name(s) found: |
SPBC17A3.05c
[Sanger Pombe]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 403 amino acids |
Gene Ontology: |
|
| Cellular Component: |
integral to membrane
[IEA]
endoplasmic reticulum [IDA |
| Biological Process: |
protein folding
[IEA]
|
| Molecular Function: |
unfolded protein binding
[IEA]
Hsp70 protein binding [ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MESSREEALR CIGLARKYLN AGEYDKALKF ANKSLRIHAT TEGEDFVKQI EKQKLTGSPN 60
61 PQATPKQENK SNFFSEKQSV RENGNSSAGE KKQKWTSEQH LLVQKIIKYK NHQYYEILDL 120
121 KKTCTDTEIK KSYKKLALQL HPDKNHAPSA DEAFKMVSKA FQVLSDPNLR AHYDRTGMDP 180
181 ESRASAASSS FSSNAGGHPG FSAYPQANMS PEDLFNSFFG DQFFSGPGTF FFGGGPGIRV 240
241 HQFGGRPRNF ARRQQAQDMP PKSIFYQLLP LIVVILFAFL SNFSWSDSTS VNTRYSFQQN 300
301 YKYTVPRTTA KHNIPYYMSQ KDLDKLSSRD IRRLNEKVEH TYTQNVHNAC LREQQIKEDE 360
361 IRRAQGWFFP DKEALKKAKE LRLPNCEELN RLGYRTYSNS YYF |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
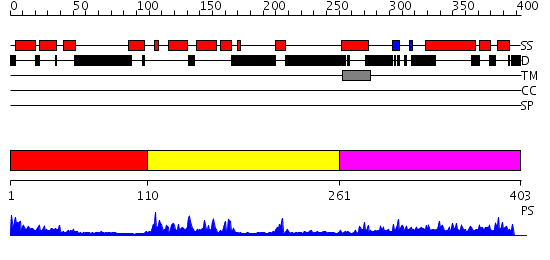
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..109] | 1.434989 | View MSA. Confident ab initio structure predictions are available. | |
| 2 | View Details | [110..260] | 18.522879 | NMR STRUCTURE OF THE J-DOMAIN (RESIDUES 2-76) IN THE ESCHERICHIA COLI N-TERMINAL FRAGMENT (RESIDUES 2-108) OF THE MOLECULAR CHAPERONE DNAJ, 20 STRUCTURES | |
| 3 | View Details | [261..403] | 2.257983 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||
| 1 | No functions predicted. | |||||||||
| 2 |
|
|||||||||
| 3 | No functions predicted. |
| Source: Reynolds et al. 2008. Manuscript submitted |

|