













| General Information: |
|
| Name(s) found: |
rga6 /
SPBC354.13
[Sanger Pombe]
|
| Description(s) found:
Found 20 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 733 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cellular_component
[ND]
|
| Biological Process: |
regulation of Rho GTPase activity
[ISS]
|
| Molecular Function: |
Rho GTPase activator activity
[ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTLNPSTLNN GTLGDLKEHK DVVLSSNAQT DEKRFEEPSL EVGGSLAKVE NGLSTSISSS 60
61 ASNLEGQQAP FFTKQNSCLS RFRELAQTYN IHEINLEEYV SQIKEVKQSL YSDDSVFNES 120
121 KSSSPPDAHT DKYFTPCGSP TKLIHSTLLE ERDTPSSLEH VSFYLQESAV SEVRFDKPSN 180
181 NGPLGRSSLN LSSLSHELQT SQDSPSLSAT NQLSSSDTLE PLQYPPSSFG SQRQFNASQD 240
241 SPKRSPSKGS WSSILRRPSL TYSPKRQSTG LHRRSLTNYF KFPHRNAHQH NAIAHNPSVF 300
301 GKPIGDLTSD PTNLCKFTFP TPECAPPSLL LPHIFAQCVH FLSLNALHVP GIFRISGSGP 360
361 VIKAITEYFY SPPHFWLDET SEIFKRIGFP SYIDIAAVLK RYIMLLPGGL FTHKDILNLL 420
421 VPLYVDSSRV KVPIDIRNEM AALCFSQIDS YVRFSILCSL LALLHQIATR TQELENGMEN 480
481 TKDSTLMKPE ALGIIFGPLL LGNSSTDLSK SCPSGNVNDL MRFETEKARV EAKIVEGLII 540
541 HWPEVLLKIN SLDIPNCFSD VGLTDQVAIF TPAVPKEDSP EQIPENVNAS EESYPNVKHI 600
601 SKLPLINDSS DNESGNQEND DAVANESTKV VVDNQQPQPK ISTVSDTAVP SMSFANNISS 660
661 RSVISAATDS KPSTRTSPPF VNNTKPIVAK SPVTVTASSE TNKKSQKINK KASPRVSLWT 720
721 KLFGKFRSNK KKS |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
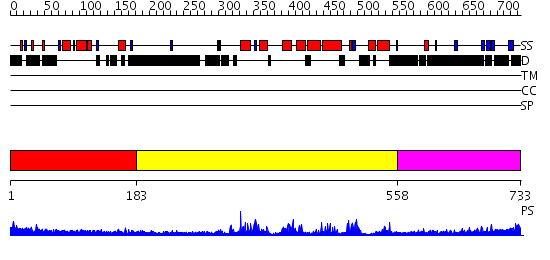
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..182] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [183..557] | 49.0 | Crystal Structure of the Human Beta2-Chimaerin | |
| 3 | View Details | [558..733] | 3.485997 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
|||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.96 |
Source: Reynolds et al. (2008)