













| General Information: |
|
| Name(s) found: |
SPCC18B5.05c
[Sanger Pombe]
|
| Description(s) found:
Found 19 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 327 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA cytosol [IDA |
| Biological Process: |
phosphorylation
[IC thiamin biosynthetic process [ISS |
| Molecular Function: |
ATP binding
[IEA]
phosphomethylpyrimidine kinase activity [ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MEANQSHNAK TNHPTCLTIA GSDCSGAAGI QADLKVMTAH QVYGMSVLTA LTCQNSHGIT 60
61 GIYPLHPSLI QRQIDACLSD IQCRVVKIGM LPDPKSIPVI SQALTKYKIT DVVMDSVIIS 120
121 SMGNVMCETP TIPATIQHLF PHLLVYASNV MEAFILVEKT LKKSPPPLKS FPDIQNLMSI 180
181 IHRLGPKFVV LRGHHVAFDK NMMITEKPDS KSWTADLIYD GKEFYIFEKP YNTTKSIHGE 240
241 SCSLTAAIAS NLACNIPPLQ AIHEALYSIE WAIQRVHKKS SDPYKSAQLF TALGHSSKFS 300
301 GSLISLKSEN YIPQADLTSV LLSHIPS |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
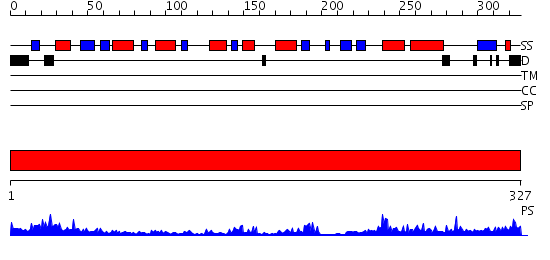
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..327] | 54.0 | 4-amino-5-hydroxymethyl-2-methylpyrimidine phosphate kinase (HMP-phosphate kinase, ThiD) |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.84 |
Source: Reynolds et al. (2008)