













| General Information: |
|
| Name(s) found: |
set2 /
SPAC29B12.02c
[Sanger Pombe]
|
| Description(s) found:
Found 19 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 798 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA nuclear chromatin [IC DNA-directed RNA polymerase II, core complex [IDA |
| Biological Process: |
histone methylation
[IDA RNA elongation from RNA polymerase II promoter [IGI regulation of transcription from RNA polymerase II promoter [IC |
| Molecular Function: |
histone methyltransferase activity (H3-K36 specific)
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MQTASSLSVL TPLNEENVDR KSSWSKDTIA VQAVGSSPSS SSSHDFESKE DAEGMNKDES 60
61 APSPSTSSPS SASSRSQSKY VRKEALPPQL FHHLDSAKDK ALTTFEEIQE CQYASANIGK 120
121 PPENEAMICD CRPHWVDGVN VACGHGSNCI NRMTSIECTD EDNVCGPSCQ NQRFQRHEFA 180
181 KVDVFLTEKK GFGLRADANL PKDTFVYEYI GEVIPEQKFR KRMRQYDSEG IKHFYFMMLQ 240
241 KGEYIDATKR GSLARFCNHS CRPNCYVDKW MVGDKLRMGI FCKRDIIRGE ELTFDYNVDR 300
301 YGAQAQPCYC GEPCCVGYIG GKTQTEAQSK LPENVREALG IEDEEDSWEN ITARRQRRKK 360
361 GIDETSKIIE EVQPTPLTSE SATKVIGVLL QTKDDLLTRK LMERIFLTSD PSVCRSIIAL 420
421 RGYNIFGLML KKFSIDIEFI LRSIKTMLSW PRLTRNKIQD SNIEPVVQEF CDHENEEVKD 480
481 HAKTLLKEWE SLEIAYRIPR RKPGQVAPQS TNAEPSNNQS NPPLRDQEPQ RGDKGDIKSA 540
541 INNSTEDLSK KHPALHSSRP SDSRSRSKFG NDYQSHSKHN LFRKNSFPKR RRLSNSDTPS 600
601 ETTTPNNEQE QVSNQANKVD LNKIISAAME SVNQKNVLKA QKEEEERIAQ QKREEKRRLA 660
661 YEESLKRHAK KLHEKKTKSS QDATIDHHLT SHSPESIAFK AVLAKFFANK TARYQEKLGK 720
721 AEFKLRVKKM TEIILKKHIQ LVLSKKEKAL PDELSDSQQR KLRVWAFRYL DTVVSRSGTA 780
781 TTTPTDSPSI GESPKKAA |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
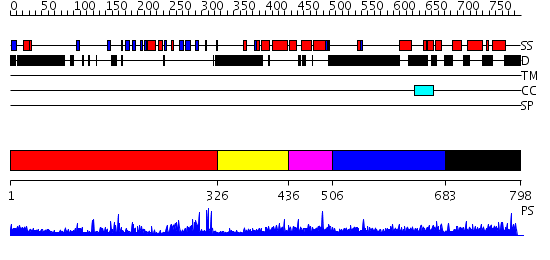
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..325] | 49.0 | SET domain of Clr4 | |
| 2 | View Details | [326..435] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [436..505] | 1.0 | Transcription elongation factor TFIIS N-domain | |
| 4 | View Details | [506..682] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [683..798] | 16.69897 | Structure and CTD binding of the Set2 SRI domain |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)