













| General Information: |
|
| Name(s) found: |
syj2 /
SPBC577.13
[Sanger Pombe]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 889 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IDA |
| Biological Process: |
cellular response to stress
[IEP phosphatidylinositol metabolic process [ISS inositol lipid-mediated signaling [IC endocytosis [ISS phosphoinositide dephosphorylation [ISS |
| Molecular Function: |
phosphatidylinositol-4,5-bisphosphate 5-phosphatase activity
[IEA]
inositol-polyphosphate 5-phosphatase activity [ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MQCYLKRKER SVAFITEKYA AIIQKPSVKD SDCIITFTCI SLEEFEKIKE YELLFGGEKI 60
61 LGTLGLFNYV CEDMKEGVFL CVARKAKLVL RIGNRDPISK LENVSFISLD QELWDEELFE 120
121 PMPRNSSPSS TFSTSTSDLN NIEKDNLVNS EYSSKYSSTT RIYPYHSLSQ LTDLLTDGSF 180
181 YVSTNISCFS RFQSEQPYGP KDIYCWNRFL ITELDNHFSE AHILEKFPNL LLHCFRGYVE 240
241 RMFIDSMSMS IISKVSSYGS RQTYPPSGID DSSYCSMFVE TEFIVEIAQT VFSFVQVRGT 300
301 VPCFWEEQFS SWYGPSISFL RSSQASQSLF NFHFSKLYKA YGDIYVIDLL QTKGFEASLY 360
361 EAYKHHLMLL PFPAVVKKFH FTPDPARPDI DPRLETDLAD DLKEMGYTQK SIENDMLESF 420
421 QKGVFRINDL DCLGRTNVIQ YQISRLVLKD IFFNLQIGVT FNILEYLRHL WSNNGDAIAK 480
481 LITGVGSIGS SATRRGRKSI AGSLSDISKS FGRMYVGRYP DVESQNAILL LLGCFPNQSP 540
541 VLISESVSSY IQGVLRQRRS EYRVERDFSI FSSTFNANGK VPSTDEFKRL LLPFGERTSA 600
601 YDLYVVAVQE IITLNMSHLV SSSNQKLRIW EEKILMILNS RDSNNKYMLI SSIQMAGVFL 660
661 GVFIRKDDHL VVSKVTKTTR KTGFGGFSAN KGAVAIEMNV CDSDFCFVSS HFAPKVNNIS 720
721 ERNMEYTSIS DNLVFPSGMK IYDHTNILWM GDFNYRIDSD NEEVRKLVEL DDLDKLASYD 780
781 QLCTEMKKGT VFHGLVEPQL TFLPTYKFDN GTNDYDTSDK QRVPSWTDRI LATKSFTRNC 840
841 YKSCDIRCSD HRPVFATFSL KIFSTCMDKK AGIIRDARIA YIKSKNSNK |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
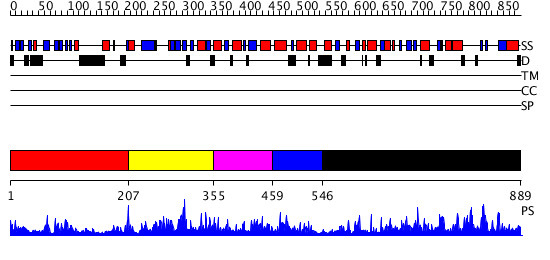
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..206] | 1.082983 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [207..354] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [355..458] | 1.091979 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [459..545] | N/A | Confident ab initio structure predictions are available. | |
| 5 | View Details | [546..889] | 113.0 | Synaptojanin, IPP5C domain |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.95 |
Source: Reynolds et al. (2008)