













| General Information: |
|
| Name(s) found: |
ero11 /
SPBC4F6.16c
[Sanger Pombe]
|
| Description(s) found:
Found 19 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 467 amino acids |
Gene Ontology: |
|
| Cellular Component: |
endoplasmic reticulum membrane
[IDA endoplasmic reticulum [IDA |
| Biological Process: |
cellular response to stress
[IEP protein folding [IC protein thiol-disulfide exchange [IGI protein folding in endoplasmic reticulum [IC] |
| Molecular Function: |
protein disulfide oxidoreductase activity
[IGI FAD binding [IEA] oxidoreductase activity, acting on sulfur group of donors, disulfide as acceptor [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MIGITKLISI WNAIWNFLLL PEIIISQTDT SSTGIYQMQS KVRSLVPVLT ERSDYFSYYR 60
61 VNLYRSSCPL WENDNAMCSN QGCAVKSLNE IEIPKVWKKL SDFEPHSKKN DTKCNWKYNP 120
121 DLDYCYLDNS TSPDEYVYVS LVQNPERFTG YAGDHSAAIW RSIYEQNCFV VDDDDNPSEQ 180
181 PKSNALFRPN QIPLNLFTEN HDDTSLSPSV ACLEKRMFNR IISGFHASIS THVCQNYYDV 240
241 EEQRWTQNLD WWRAKVGSFP DRIENIFFNY ALLHQALVQI ATQMKNITSD SSFTFCPTDK 300
301 DVDWRTHTAF EQLVYHAYQD RHVINREQFF ADEEAKRFKD SFRKHFRDIS RIMDCVGCDK 360
361 CRLWGKVQIT GYGTALKLLL EPETQLSDLR PEEVVALVNT FDRISHSVEA TFWFSLKAKT 420
421 NDTLDAIKAL IYKTYRSPSK EILAFFIDQT WYLFYALFFI CNVPRVI |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
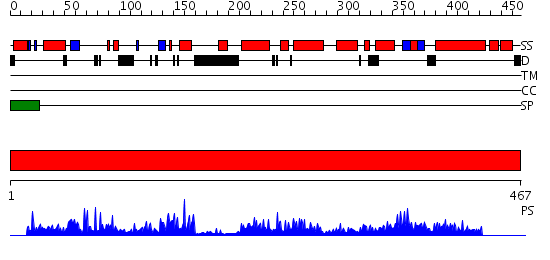
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..467] | 111.0 | Structure of Ero1p, Source of Disulfide Bonds for Oxidative Protein Folding in the Cell |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.45 |
Source: Reynolds et al. (2008)